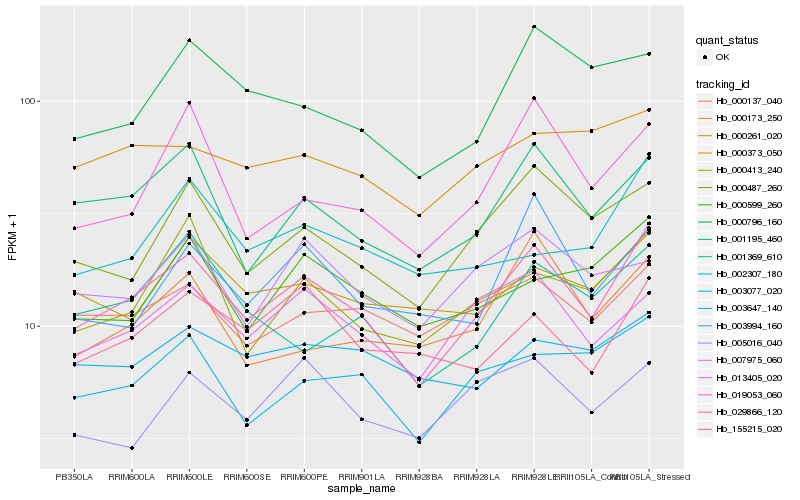
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029866_120 |
0.0 |
- |
- |
hypothetical protein CICLE_v10006070mg [Citrus clementina] |
| 2 |
Hb_001195_460 |
0.0958789715 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000487_260 |
0.1002416278 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 4 |
Hb_013405_020 |
0.1036118242 |
- |
- |
- |
| 5 |
Hb_003077_020 |
0.1064546131 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_000413_240 |
0.1111251745 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000137_040 |
0.1127424511 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 8 |
Hb_155215_020 |
0.1147804257 |
- |
- |
PREDICTED: RNA-binding protein pno1-like [Populus euphratica] |
| 9 |
Hb_000599_260 |
0.1161176882 |
- |
- |
Vacuolar protein sorting-associated protein 2 like 3 [Glycine soja] |
| 10 |
Hb_003994_160 |
0.1161450985 |
- |
- |
PREDICTED: uncharacterized protein LOC105131691 isoform X5 [Populus euphratica] |
| 11 |
Hb_019053_060 |
0.1164342975 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 12 |
Hb_000373_050 |
0.1166908372 |
- |
- |
PREDICTED: protein WHI3 [Jatropha curcas] |
| 13 |
Hb_000796_160 |
0.119547892 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001369_610 |
0.1207868404 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000261_020 |
0.1218341987 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003647_140 |
0.1233210264 |
- |
- |
PREDICTED: uncharacterized protein LOC105646673 [Jatropha curcas] |
| 17 |
Hb_000173_250 |
0.1236510064 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase small chain A [Jatropha curcas] |
| 18 |
Hb_005016_040 |
0.1238431059 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X2 [Jatropha curcas] |
| 19 |
Hb_002307_180 |
0.1242868628 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_007975_060 |
0.1261112032 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |