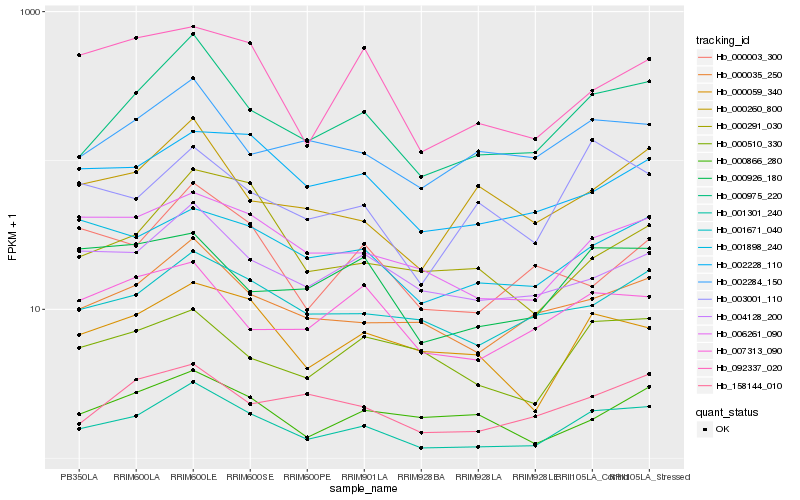
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001301_240 |
0.0 |
- |
- |
- |
| 2 |
Hb_000975_220 |
0.1013842419 |
- |
- |
Histone H2B [Medicago truncatula] |
| 3 |
Hb_000035_250 |
0.1497024119 |
- |
- |
PREDICTED: uncharacterized protein LOC105637544 [Jatropha curcas] |
| 4 |
Hb_000059_340 |
0.1517273632 |
transcription factor |
TF Family: SOH1 |
uncharacterized protein LOC100527879 [Glycine max] |
| 5 |
Hb_006261_090 |
0.1561619899 |
- |
- |
PREDICTED: exosome complex exonuclease RRP46 homolog [Jatropha curcas] |
| 6 |
Hb_158144_010 |
0.1645330387 |
- |
- |
- |
| 7 |
Hb_001671_040 |
0.1662122971 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS31-like isoform X1 [Populus euphratica] |
| 8 |
Hb_004128_200 |
0.1699559589 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 9 |
Hb_000926_180 |
0.171266482 |
- |
- |
proline synthetase associated protein, putative [Ricinus communis] |
| 10 |
Hb_000510_330 |
0.1726917757 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 11 |
Hb_001898_240 |
0.1728198972 |
- |
- |
PREDICTED: uncharacterized protein LOC105779688 [Gossypium raimondii] |
| 12 |
Hb_092337_020 |
0.1728410676 |
- |
- |
hypothetical protein B456_013G264900 [Gossypium raimondii] |
| 13 |
Hb_000260_800 |
0.1730365248 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 [Populus euphratica] |
| 14 |
Hb_000291_030 |
0.1732936574 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 15 |
Hb_002228_110 |
0.1736819509 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 16 |
Hb_003001_110 |
0.1750619582 |
- |
- |
oleoyl-acyl carrier protein thioesterase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000866_280 |
0.1753881621 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 18 |
Hb_007313_090 |
0.179008215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000003_300 |
0.1791363131 |
- |
- |
PREDICTED: uncharacterized protein LOC105631182 [Jatropha curcas] |
| 20 |
Hb_002284_150 |
0.179138371 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |