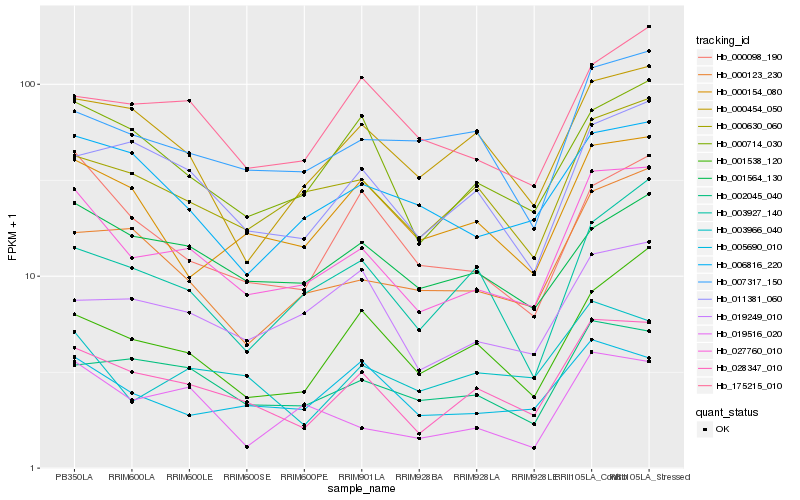
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027760_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630460 [Jatropha curcas] |
| 2 |
Hb_028347_010 |
0.0892855471 |
- |
- |
- |
| 3 |
Hb_000630_060 |
0.1054547083 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 1, mitochondrial [Jatropha curcas] |
| 4 |
Hb_002045_040 |
0.1190938875 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000714_030 |
0.1210192709 |
- |
- |
Nucleosome assembly [Gossypium arboreum] |
| 6 |
Hb_000123_230 |
0.121963931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_019516_020 |
0.1260234604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006816_220 |
0.1283402474 |
- |
- |
PREDICTED: zinc finger matrin-type protein 2 [Jatropha curcas] |
| 9 |
Hb_007317_150 |
0.1295804543 |
- |
- |
PREDICTED: 40S ribosomal protein S25 [Jatropha curcas] |
| 10 |
Hb_000098_190 |
0.1302209228 |
- |
- |
PREDICTED: uncharacterized protein LOC105633343 [Jatropha curcas] |
| 11 |
Hb_011381_060 |
0.1309435434 |
- |
- |
PREDICTED: ADP-ribosylation factor 3 [Jatropha curcas] |
| 12 |
Hb_000154_080 |
0.1321272891 |
transcription factor |
TF Family: bZIP |
PREDICTED: probable transcription factor PosF21 [Jatropha curcas] |
| 13 |
Hb_175215_010 |
0.1335924476 |
- |
- |
hypothetical protein CISIN_1g029605mg [Citrus sinensis] |
| 14 |
Hb_005690_010 |
0.1341525333 |
- |
- |
PREDICTED: origin recognition complex subunit 2 [Populus euphratica] |
| 15 |
Hb_001538_120 |
0.1341724884 |
- |
- |
PREDICTED: FAD-linked sulfhydryl oxidase ERV1 [Jatropha curcas] |
| 16 |
Hb_001564_130 |
0.1356720651 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_003966_040 |
0.1363484836 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 18 |
Hb_000454_050 |
0.1375954023 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 [Vitis vinifera] |
| 19 |
Hb_003927_140 |
0.138014166 |
- |
- |
PREDICTED: nudix hydrolase 9 isoform X1 [Jatropha curcas] |
| 20 |
Hb_019249_010 |
0.1385990687 |
- |
- |
PREDICTED: origin of replication complex subunit 2 [Jatropha curcas] |