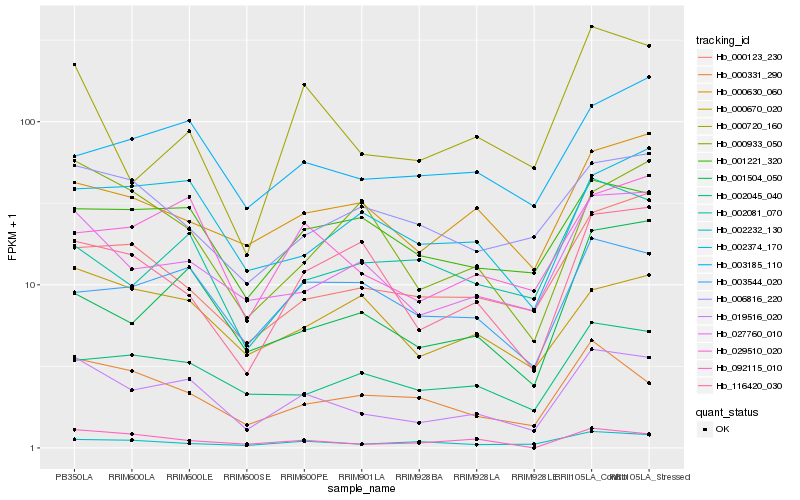
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019516_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_027760_010 |
0.1260234604 |
- |
- |
PREDICTED: uncharacterized protein LOC105630460 [Jatropha curcas] |
| 3 |
Hb_029510_020 |
0.1433871428 |
- |
- |
hypothetical protein JCGZ_06306 [Jatropha curcas] |
| 4 |
Hb_002045_040 |
0.1570754114 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001221_320 |
0.1585229582 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000331_290 |
0.1596263962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002374_170 |
0.1596610728 |
- |
- |
20S proteasome beta subunit C1 [Hevea brasiliensis] |
| 8 |
Hb_001504_050 |
0.1647405632 |
- |
- |
hypothetical protein POPTR_0001s14840g [Populus trichocarpa] |
| 9 |
Hb_000630_060 |
0.1655749748 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 1, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000720_160 |
0.1656044029 |
- |
- |
PREDICTED: C2 domain-containing protein At1g63220 [Jatropha curcas] |
| 11 |
Hb_092115_010 |
0.1657789536 |
- |
- |
- |
| 12 |
Hb_000123_230 |
0.1665792171 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003544_020 |
0.1670554346 |
- |
- |
PREDICTED: elongin-A [Jatropha curcas] |
| 14 |
Hb_002232_130 |
0.1671026476 |
- |
- |
- |
| 15 |
Hb_000670_020 |
0.1673994942 |
- |
- |
PREDICTED: chromatin modification-related protein EAF3-like [Jatropha curcas] |
| 16 |
Hb_116420_030 |
0.1703312923 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 17 |
Hb_006816_220 |
0.1714405522 |
- |
- |
PREDICTED: zinc finger matrin-type protein 2 [Jatropha curcas] |
| 18 |
Hb_000933_050 |
0.1732922921 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_002081_070 |
0.1738798893 |
- |
- |
PREDICTED: pyridoxal kinase [Jatropha curcas] |
| 20 |
Hb_003185_110 |
0.1741792643 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 7-2 [Nelumbo nucifera] |