| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
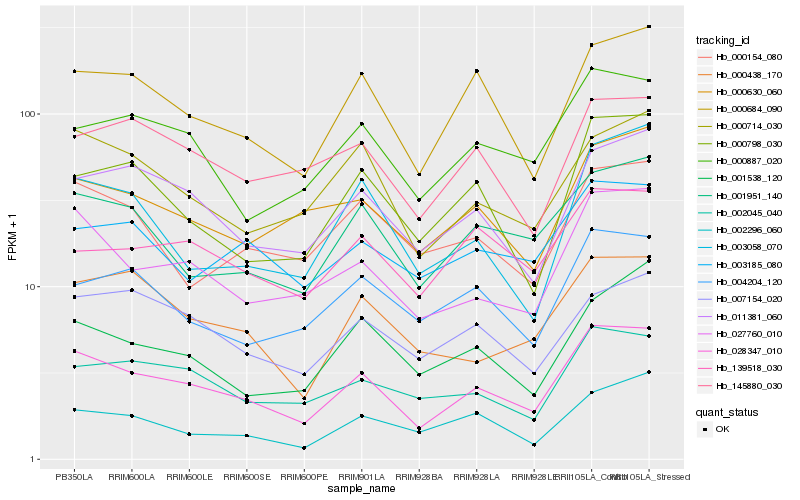
Hb_028347_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_027760_010 |
0.0892855471 |
- |
- |
PREDICTED: uncharacterized protein LOC105630460 [Jatropha curcas] |
| 3 |
Hb_000684_090 |
0.0990624847 |
transcription factor |
TF Family: S1Fa-like |
- |
| 4 |
Hb_011381_060 |
0.1035970573 |
- |
- |
PREDICTED: ADP-ribosylation factor 3 [Jatropha curcas] |
| 5 |
Hb_001951_140 |
0.1058748375 |
- |
- |
PREDICTED: uncharacterized protein LOC105649757 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000887_020 |
0.1090720734 |
- |
- |
PREDICTED: uncharacterized protein LOC105633527 [Jatropha curcas] |
| 7 |
Hb_002045_040 |
0.1113682958 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000714_030 |
0.1132015903 |
- |
- |
Nucleosome assembly [Gossypium arboreum] |
| 9 |
Hb_000798_030 |
0.114620273 |
- |
- |
PREDICTED: uncharacterized protein LOC105635919 [Jatropha curcas] |
| 10 |
Hb_000630_060 |
0.1159792944 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 1, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000438_170 |
0.1200425797 |
- |
- |
- |
| 12 |
Hb_004204_120 |
0.1227702982 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 7 [Jatropha curcas] |
| 13 |
Hb_002296_060 |
0.1238392319 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Fragaria vesca subsp. vesca] |
| 14 |
Hb_003058_070 |
0.1241207105 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 15 |
Hb_000154_080 |
0.1248809784 |
transcription factor |
TF Family: bZIP |
PREDICTED: probable transcription factor PosF21 [Jatropha curcas] |
| 16 |
Hb_145880_030 |
0.1249605002 |
- |
- |
ubiquitin-conjugating enzyme E2-25kD, putative [Ricinus communis] |
| 17 |
Hb_139518_030 |
0.1258124719 |
- |
- |
protein kinase atn1, putative [Ricinus communis] |
| 18 |
Hb_007154_020 |
0.1267154943 |
- |
- |
- |
| 19 |
Hb_003185_080 |
0.126845033 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 20 |
Hb_001538_120 |
0.1270172119 |
- |
- |
PREDICTED: FAD-linked sulfhydryl oxidase ERV1 [Jatropha curcas] |