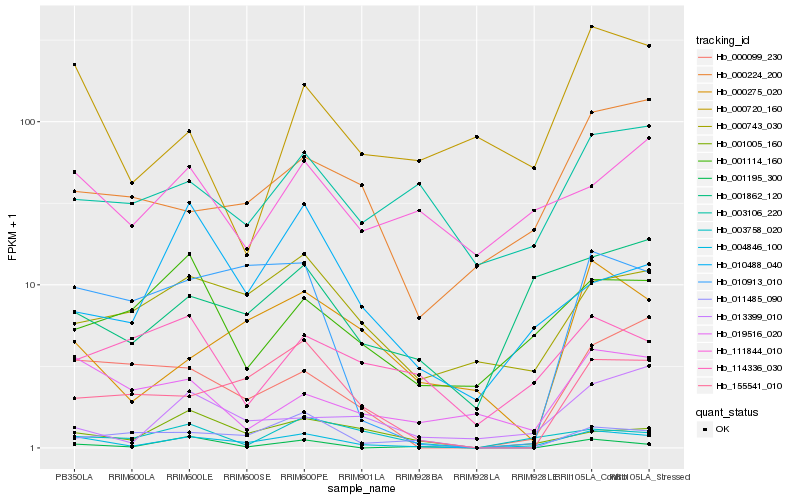
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004846_100 |
0.0 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 2 |
Hb_155541_010 |
0.2346313684 |
- |
- |
- |
| 3 |
Hb_001862_120 |
0.2374190729 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 [Jatropha curcas] |
| 4 |
Hb_000743_030 |
0.2384144978 |
- |
- |
NmrA-like negative transcriptional regulator family protein [Theobroma cacao] |
| 5 |
Hb_010913_010 |
0.2425946532 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 6 |
Hb_000275_020 |
0.2453630109 |
- |
- |
PREDICTED: uncharacterized protein LOC105632656 [Jatropha curcas] |
| 7 |
Hb_001005_160 |
0.2459461652 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000099_230 |
0.2517152742 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Gossypium raimondii] |
| 9 |
Hb_003106_220 |
0.2569504285 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000224_200 |
0.2577092815 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 11 |
Hb_019516_020 |
0.2590435994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003758_020 |
0.2608872498 |
- |
- |
UGTPg34 [Panax ginseng] |
| 13 |
Hb_001114_160 |
0.2627187002 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000720_160 |
0.2651343754 |
- |
- |
PREDICTED: C2 domain-containing protein At1g63220 [Jatropha curcas] |
| 15 |
Hb_013399_010 |
0.2658615277 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 16 |
Hb_011485_090 |
0.2683265726 |
- |
- |
- |
| 17 |
Hb_111844_010 |
0.2691976676 |
- |
- |
PREDICTED: uncharacterized protein LOC105632109 [Jatropha curcas] |
| 18 |
Hb_001195_300 |
0.2706228691 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 19 |
Hb_010488_040 |
0.2732839554 |
- |
- |
PREDICTED: phosphomannomutase [Populus euphratica] |
| 20 |
Hb_114336_030 |
0.2739437782 |
- |
- |
PREDICTED: uncharacterized protein LOC105642104 [Jatropha curcas] |