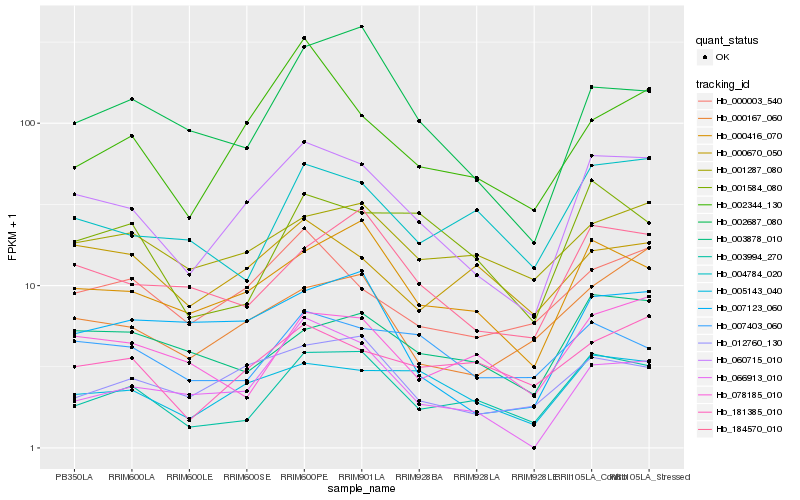
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_060715_010 |
0.0 |
- |
- |
PREDICTED: notchless protein homolog [Gossypium raimondii] |
| 2 |
Hb_005143_040 |
0.1271125996 |
- |
- |
hypothetical protein JCGZ_17315 [Jatropha curcas] |
| 3 |
Hb_000416_070 |
0.1382834507 |
- |
- |
hypothetical protein JCGZ_21620 [Jatropha curcas] |
| 4 |
Hb_000003_540 |
0.1413437553 |
- |
- |
PREDICTED: homeobox-leucine zipper protein ATHB-15 [Jatropha curcas] |
| 5 |
Hb_012760_130 |
0.1495920061 |
- |
- |
PREDICTED: uncharacterized protein LOC105647093 [Jatropha curcas] |
| 6 |
Hb_184570_010 |
0.1508696644 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 7 |
Hb_003994_270 |
0.1522715698 |
- |
- |
PREDICTED: uncharacterized protein LOC105646152 [Jatropha curcas] |
| 8 |
Hb_002344_130 |
0.1586776233 |
- |
- |
PREDICTED: copper transporter 5 [Jatropha curcas] |
| 9 |
Hb_000167_060 |
0.158708014 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 31 kDa protein [Jatropha curcas] |
| 10 |
Hb_181385_010 |
0.161263791 |
- |
- |
PREDICTED: uncharacterized protein LOC105650319 [Jatropha curcas] |
| 11 |
Hb_007123_060 |
0.1615594067 |
- |
- |
PREDICTED: uncharacterized protein LOC105634060 [Jatropha curcas] |
| 12 |
Hb_003878_010 |
0.1644873359 |
- |
- |
PREDICTED: uncharacterized protein LOC105632145 [Jatropha curcas] |
| 13 |
Hb_066913_010 |
0.1654802716 |
- |
- |
hypothetical protein POPTR_0006s03150g [Populus trichocarpa] |
| 14 |
Hb_001287_080 |
0.1655906451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007403_060 |
0.1697046345 |
- |
- |
PREDICTED: CAAX prenyl protease 2 [Jatropha curcas] |
| 16 |
Hb_078185_010 |
0.1697120572 |
- |
- |
- |
| 17 |
Hb_002687_080 |
0.1714785024 |
- |
- |
hypothetical protein JCGZ_06535 [Jatropha curcas] |
| 18 |
Hb_001584_080 |
0.1719701119 |
- |
- |
hypothetical protein JCGZ_23206 [Jatropha curcas] |
| 19 |
Hb_004784_020 |
0.172173537 |
- |
- |
PREDICTED: uncharacterized protein LOC105636452 [Jatropha curcas] |
| 20 |
Hb_000670_050 |
0.1724640113 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_16088 [Jatropha curcas] |