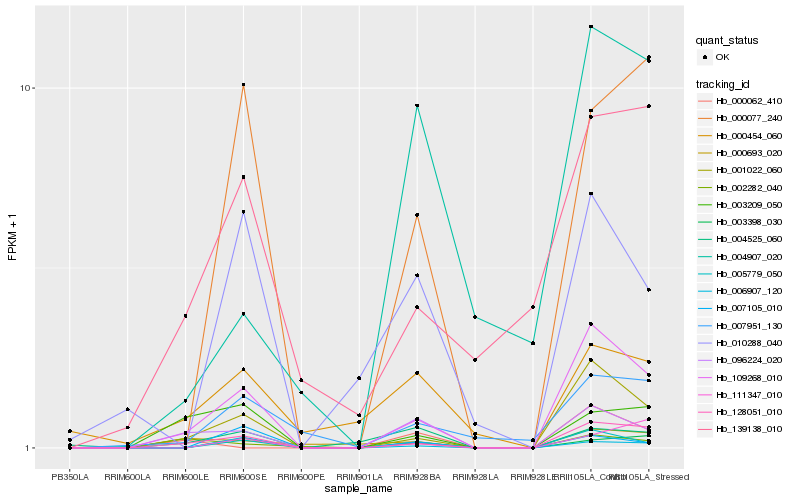
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_109268_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 2 |
Hb_001022_060 |
0.0729049607 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 3 |
Hb_128051_010 |
0.0975008884 |
- |
- |
- |
| 4 |
Hb_005779_050 |
0.2012988555 |
- |
- |
hypothetical protein JCGZ_14616 [Jatropha curcas] |
| 5 |
Hb_096224_020 |
0.2497502681 |
- |
- |
hypothetical protein JCGZ_06980 [Jatropha curcas] |
| 6 |
Hb_000077_240 |
0.2586171876 |
- |
- |
hypothetical protein PHAVU_011G114500g, partial [Phaseolus vulgaris] |
| 7 |
Hb_003398_030 |
0.2737298887 |
- |
- |
PREDICTED: uncharacterized protein LOC104583892 [Brachypodium distachyon] |
| 8 |
Hb_003209_050 |
0.2800200836 |
- |
- |
hypothetical protein JCGZ_15683 [Jatropha curcas] |
| 9 |
Hb_007951_130 |
0.2842722168 |
desease resistance |
Gene Name: UBN2_3 |
JHL25H03.6 [Jatropha curcas] |
| 10 |
Hb_006907_120 |
0.2858941394 |
- |
- |
PREDICTED: uncharacterized protein LOC103327233 [Prunus mume] |
| 11 |
Hb_139138_010 |
0.2884780857 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 12 |
Hb_111347_010 |
0.3095321519 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 13 |
Hb_010288_040 |
0.3134623706 |
- |
- |
PREDICTED: transmembrane protein 205-like [Eucalyptus grandis] |
| 14 |
Hb_002282_040 |
0.3135369549 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 15 |
Hb_000693_020 |
0.3184926688 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 16 |
Hb_000454_060 |
0.3299143681 |
- |
- |
Uncharacterized protein TCM_006778 [Theobroma cacao] |
| 17 |
Hb_004907_020 |
0.3347683083 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 18 |
Hb_007105_010 |
0.3351022216 |
- |
- |
PREDICTED: uncharacterized protein LOC103455579 [Malus domestica] |
| 19 |
Hb_000062_410 |
0.3368629477 |
- |
- |
PREDICTED: uncharacterized protein LOC105644766 [Jatropha curcas] |
| 20 |
Hb_004525_060 |
0.3379645221 |
- |
- |
- |