| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
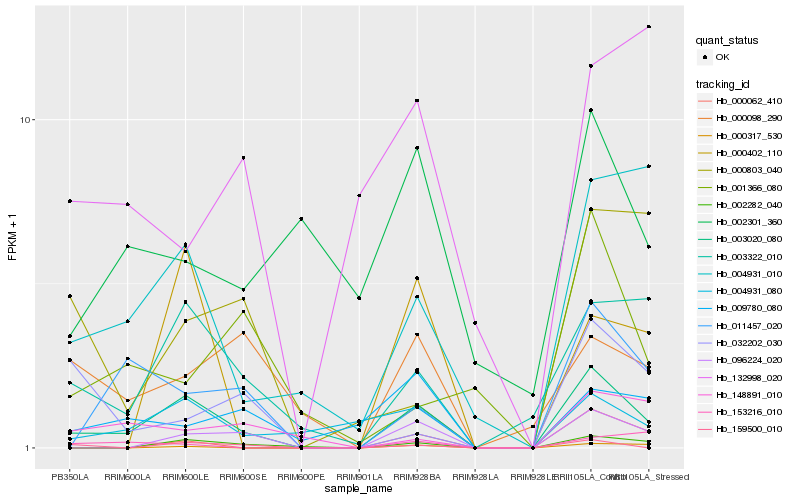
Hb_003020_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_003322_010 |
0.269643383 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_096224_020 |
0.2837478318 |
- |
- |
hypothetical protein JCGZ_06980 [Jatropha curcas] |
| 4 |
Hb_002282_040 |
0.2885586258 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 5 |
Hb_004931_010 |
0.2930877955 |
- |
- |
inosine triphosphate pyrophosphatase, putative [Ricinus communis] |
| 6 |
Hb_004931_080 |
0.3059525965 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Jatropha curcas] |
| 7 |
Hb_011457_020 |
0.3107273589 |
- |
- |
PREDICTED: uncharacterized protein LOC105649960 [Jatropha curcas] |
| 8 |
Hb_159500_010 |
0.326981017 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 9 |
Hb_000098_290 |
0.3340032967 |
- |
- |
- |
| 10 |
Hb_009780_080 |
0.3433522495 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X2 [Jatropha curcas] |
| 11 |
Hb_148891_010 |
0.3484406449 |
- |
- |
- |
| 12 |
Hb_000062_410 |
0.3533682579 |
- |
- |
PREDICTED: uncharacterized protein LOC105644766 [Jatropha curcas] |
| 13 |
Hb_000317_530 |
0.3550750827 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 14 |
Hb_000803_040 |
0.36421225 |
- |
- |
- |
| 15 |
Hb_000402_110 |
0.3644360988 |
- |
- |
hypothetical protein JCGZ_17786 [Jatropha curcas] |
| 16 |
Hb_002301_360 |
0.3665234689 |
- |
- |
hypothetical protein CICLE_v10017252mg [Citrus clementina] |
| 17 |
Hb_153216_010 |
0.3667608492 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 18 |
Hb_032202_030 |
0.3678472432 |
- |
- |
- |
| 19 |
Hb_001366_080 |
0.3712444251 |
transcription factor |
TF Family: NAC |
NAC transcription factor 068 [Jatropha curcas] |
| 20 |
Hb_132998_020 |
0.3802366294 |
- |
- |
acyl carrier protein 1, chloroplastic-like [Jatropha curcas] |