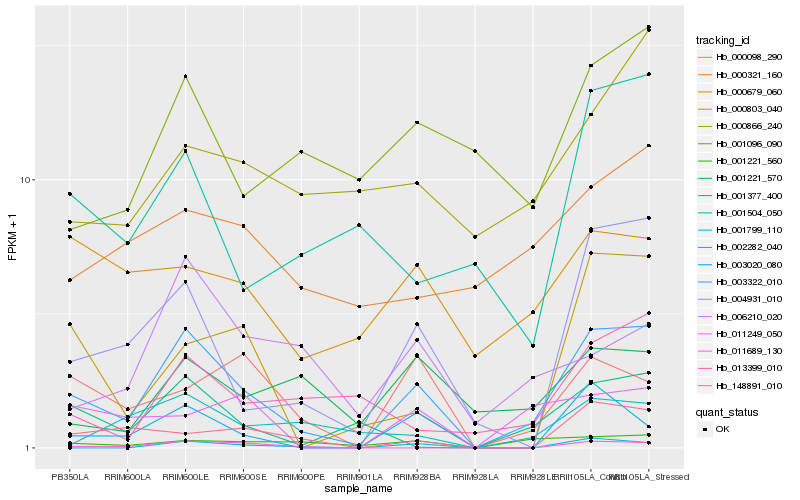
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003322_010 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_004931_010 |
0.2285619799 |
- |
- |
inosine triphosphate pyrophosphatase, putative [Ricinus communis] |
| 3 |
Hb_001221_560 |
0.2660008045 |
- |
- |
PREDICTED: 11-beta-hydroxysteroid dehydrogenase 1B-like [Jatropha curcas] |
| 4 |
Hb_003020_080 |
0.269643383 |
- |
- |
- |
| 5 |
Hb_000098_290 |
0.2715836353 |
- |
- |
- |
| 6 |
Hb_000803_040 |
0.2771814965 |
- |
- |
- |
| 7 |
Hb_001377_400 |
0.2777366628 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 8 |
Hb_148891_010 |
0.2812101596 |
- |
- |
- |
| 9 |
Hb_011249_050 |
0.28451869 |
- |
- |
- |
| 10 |
Hb_001504_050 |
0.2850733836 |
- |
- |
hypothetical protein POPTR_0001s14840g [Populus trichocarpa] |
| 11 |
Hb_013399_010 |
0.2865520447 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 12 |
Hb_011689_130 |
0.2887585402 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001221_570 |
0.2905112214 |
- |
- |
- |
| 14 |
Hb_006210_020 |
0.2908009128 |
- |
- |
PREDICTED: calcium sensing receptor, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001799_110 |
0.2931675079 |
- |
- |
PREDICTED: glycine--tRNA ligase 2, chloroplastic/mitochondrial isoform X2 [Jatropha curcas] |
| 16 |
Hb_002282_040 |
0.2935012424 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 17 |
Hb_000679_060 |
0.2945041535 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 18 |
Hb_000321_160 |
0.2959176657 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001096_090 |
0.2979445379 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000866_240 |
0.2985410622 |
- |
- |
PREDICTED: 60S ribosomal protein L36-2-like [Jatropha curcas] |