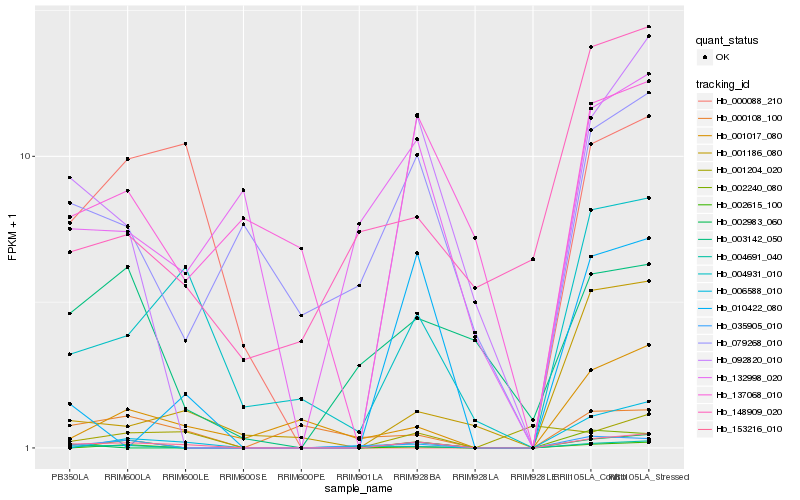
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_153216_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 2 |
Hb_004931_010 |
0.2338596282 |
- |
- |
inosine triphosphate pyrophosphatase, putative [Ricinus communis] |
| 3 |
Hb_092820_010 |
0.2552505684 |
- |
- |
hypothetical protein CISIN_1g033834mg [Citrus sinensis] |
| 4 |
Hb_006588_010 |
0.2825556632 |
- |
- |
- |
| 5 |
Hb_004691_040 |
0.3037717757 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 6 |
Hb_001017_080 |
0.3046750984 |
- |
- |
hypothetical protein POPTR_0010s06020g [Populus trichocarpa] |
| 7 |
Hb_035905_010 |
0.3056900442 |
- |
- |
PREDICTED: uncharacterized protein LOC105627956 [Jatropha curcas] |
| 8 |
Hb_001186_080 |
0.3058314578 |
- |
- |
PREDICTED: F-box protein FBW2-like [Nelumbo nucifera] |
| 9 |
Hb_010422_080 |
0.3109171924 |
- |
- |
- |
| 10 |
Hb_002240_080 |
0.3238965154 |
- |
- |
- |
| 11 |
Hb_002615_100 |
0.3252675639 |
- |
- |
- |
| 12 |
Hb_002983_060 |
0.3270666169 |
desease resistance |
Gene Name: NB-ARC |
Cc-nbs-lrr resistance protein, putative [Theobroma cacao] |
| 13 |
Hb_137068_010 |
0.3294949968 |
- |
- |
hypothetical protein CISIN_1g003571mg [Citrus sinensis] |
| 14 |
Hb_132998_020 |
0.3318925449 |
- |
- |
acyl carrier protein 1, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_079268_010 |
0.3337209912 |
- |
- |
- |
| 16 |
Hb_000108_100 |
0.333968314 |
- |
- |
PREDICTED: DNA polymerase epsilon subunit 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001204_020 |
0.3351166484 |
- |
- |
Uncharacterized protein TCM_021965 [Theobroma cacao] |
| 18 |
Hb_148909_020 |
0.3395739416 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 19 |
Hb_000088_210 |
0.3408057523 |
- |
- |
hypothetical protein POPTR_0013s11310g [Populus trichocarpa] |
| 20 |
Hb_003142_050 |
0.3410594712 |
- |
- |
- |