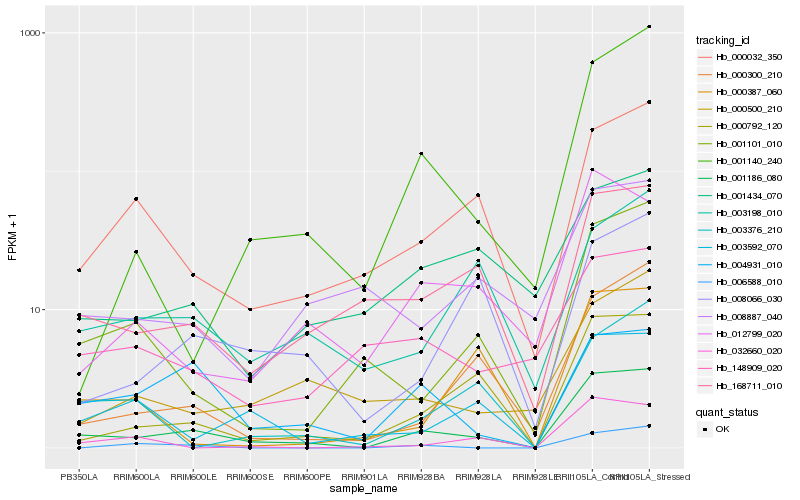
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001186_080 |
0.0 |
- |
- |
PREDICTED: F-box protein FBW2-like [Nelumbo nucifera] |
| 2 |
Hb_000792_120 |
0.198390502 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_168711_010 |
0.2138172311 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 [Sesamum indicum] |
| 4 |
Hb_000032_350 |
0.2179629782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000300_210 |
0.2194512331 |
- |
- |
gd2b, putative [Ricinus communis] |
| 6 |
Hb_001101_010 |
0.2221696943 |
- |
- |
hypothetical protein POPTR_0004s18730g [Populus trichocarpa] |
| 7 |
Hb_004931_010 |
0.2240954032 |
- |
- |
inosine triphosphate pyrophosphatase, putative [Ricinus communis] |
| 8 |
Hb_003592_070 |
0.2267378765 |
- |
- |
- |
| 9 |
Hb_012799_020 |
0.2361392236 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 10 |
Hb_001140_240 |
0.2389502332 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 11 |
Hb_000500_210 |
0.2397638734 |
- |
- |
- |
| 12 |
Hb_003376_210 |
0.2437870182 |
- |
- |
- |
| 13 |
Hb_003198_010 |
0.2449776529 |
- |
- |
Valine--tRNA ligase [Gossypium arboreum] |
| 14 |
Hb_008066_030 |
0.2453546004 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 15 |
Hb_032660_020 |
0.2476239188 |
- |
- |
PREDICTED: F-box protein At5g49610-like [Jatropha curcas] |
| 16 |
Hb_001434_070 |
0.2492833232 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM8 [Jatropha curcas] |
| 17 |
Hb_006588_010 |
0.2500379326 |
- |
- |
- |
| 18 |
Hb_148909_020 |
0.2509226236 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 19 |
Hb_008887_040 |
0.2565480425 |
- |
- |
Ribonuclease P family protein / Rpp14 family protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000387_060 |
0.2583482125 |
- |
- |
- |