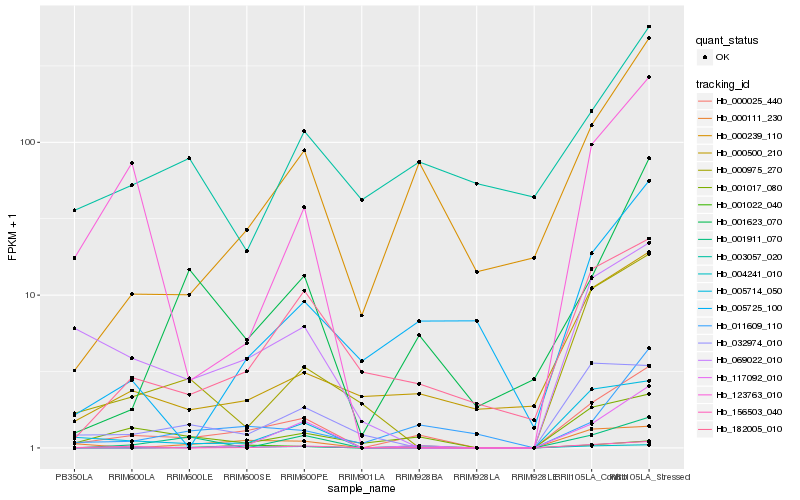
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_440 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein P16F5.05c-like [Populus euphratica] |
| 2 |
Hb_001017_080 |
0.2061226556 |
- |
- |
hypothetical protein POPTR_0010s06020g [Populus trichocarpa] |
| 3 |
Hb_182005_010 |
0.2206684515 |
- |
- |
PREDICTED: uncharacterized protein LOC105639166 [Jatropha curcas] |
| 4 |
Hb_000239_110 |
0.221494353 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 5 |
Hb_156503_040 |
0.2243927079 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 6 |
Hb_001022_040 |
0.2304412085 |
- |
- |
PREDICTED: uncharacterized protein LOC105634753 [Jatropha curcas] |
| 7 |
Hb_000500_210 |
0.23478454 |
- |
- |
- |
| 8 |
Hb_005714_050 |
0.2361535498 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_1461s00200g [Populus trichocarpa] |
| 9 |
Hb_069022_010 |
0.2393543703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001623_070 |
0.2499319559 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 11 |
Hb_000975_270 |
0.2501296955 |
- |
- |
PREDICTED: histone H2B.11-like [Phoenix dactylifera] |
| 12 |
Hb_011609_110 |
0.2525650794 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 13 |
Hb_001911_070 |
0.2628189921 |
- |
- |
- |
| 14 |
Hb_004241_010 |
0.2657136046 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 15 |
Hb_032974_010 |
0.2701250895 |
- |
- |
- |
| 16 |
Hb_117092_010 |
0.2705181165 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 17 |
Hb_005725_100 |
0.272262923 |
- |
- |
hypothetical protein 30 [Hevea brasiliensis] |
| 18 |
Hb_123763_010 |
0.2788377073 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 19 |
Hb_000111_230 |
0.2790298145 |
- |
- |
- |
| 20 |
Hb_003057_020 |
0.2795138116 |
- |
- |
40S ribosomal protein S23, putative [Ricinus communis] |