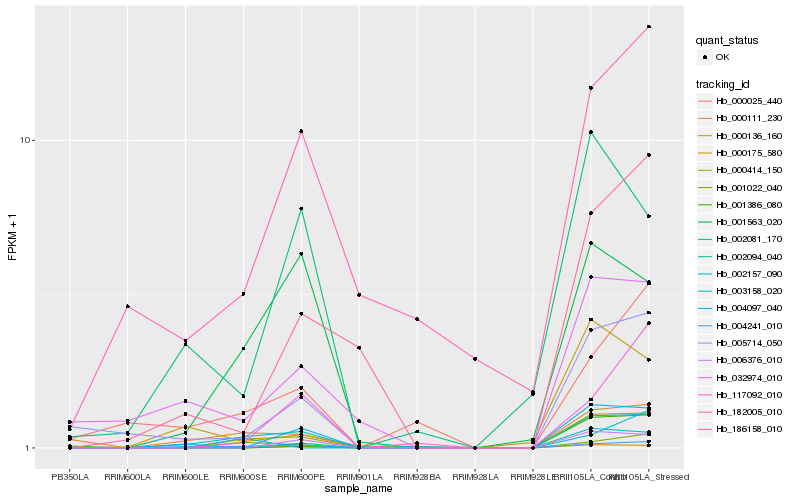
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000414_150 |
0.0 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 2 |
Hb_004241_010 |
0.2358107631 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 3 |
Hb_002094_040 |
0.2408616702 |
- |
- |
Organic cation transporter protein [Aegilops tauschii] |
| 4 |
Hb_006376_010 |
0.2449534249 |
- |
- |
PREDICTED: uncharacterized protein LOC104090718 [Nicotiana tomentosiformis] |
| 5 |
Hb_001563_020 |
0.2470568659 |
transcription factor |
TF Family: TRAF |
aberrant large forked product, putative [Ricinus communis] |
| 6 |
Hb_000111_230 |
0.2475251636 |
- |
- |
- |
| 7 |
Hb_005714_050 |
0.2534873748 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_1461s00200g [Populus trichocarpa] |
| 8 |
Hb_001022_040 |
0.2569147115 |
- |
- |
PREDICTED: uncharacterized protein LOC105634753 [Jatropha curcas] |
| 9 |
Hb_002157_090 |
0.2624021201 |
- |
- |
hypothetical protein VITISV_041741 [Vitis vinifera] |
| 10 |
Hb_000136_160 |
0.2654678842 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 11 |
Hb_004097_040 |
0.2685606348 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_117092_010 |
0.2852358619 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 13 |
Hb_186158_010 |
0.2913852409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_032974_010 |
0.2986562863 |
- |
- |
- |
| 15 |
Hb_001386_080 |
0.3053025095 |
- |
- |
hypothetical protein JCGZ_06052 [Jatropha curcas] |
| 16 |
Hb_000175_580 |
0.3058883047 |
transcription factor, desease resistance |
TF Family: C3H, Gene Name: zf-CCCH |
hypothetical protein POPTR_0013s00890g [Populus trichocarpa] |
| 17 |
Hb_000025_440 |
0.3073084016 |
- |
- |
PREDICTED: ankyrin repeat-containing protein P16F5.05c-like [Populus euphratica] |
| 18 |
Hb_182005_010 |
0.3145082581 |
- |
- |
PREDICTED: uncharacterized protein LOC105639166 [Jatropha curcas] |
| 19 |
Hb_002081_170 |
0.3157177089 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 20 |
Hb_003158_020 |
0.3220965294 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |