| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
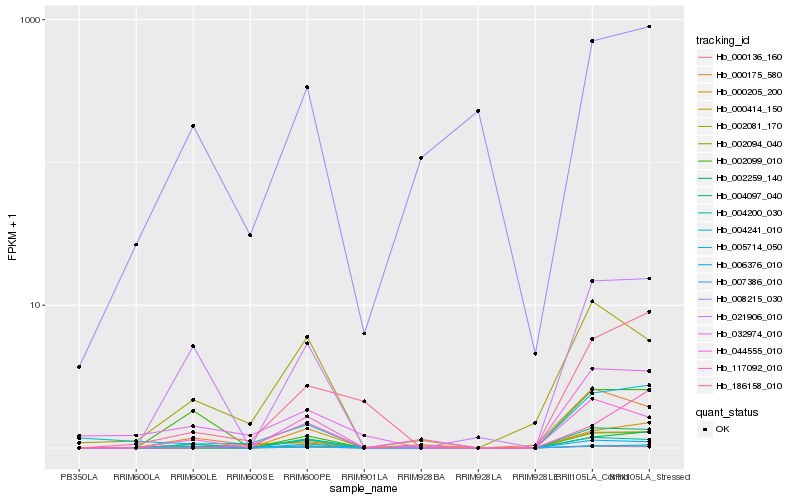
Hb_004097_040 |
0.0 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_006376_010 |
0.1521603295 |
- |
- |
PREDICTED: uncharacterized protein LOC104090718 [Nicotiana tomentosiformis] |
| 3 |
Hb_002259_140 |
0.1683427288 |
- |
- |
- |
| 4 |
Hb_000136_160 |
0.1694976775 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 5 |
Hb_021906_010 |
0.170957034 |
- |
- |
PREDICTED: cysteine proteinase inhibitor-like [Populus euphratica] |
| 6 |
Hb_044555_010 |
0.1740730455 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 7 |
Hb_007386_010 |
0.1840140026 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 8 |
Hb_004200_030 |
0.1918734793 |
- |
- |
PREDICTED: SWR1 complex subunit 6-like isoform X1 [Citrus sinensis] |
| 9 |
Hb_002094_040 |
0.2146909988 |
- |
- |
Organic cation transporter protein [Aegilops tauschii] |
| 10 |
Hb_002081_170 |
0.2209660833 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 11 |
Hb_000205_200 |
0.2210287533 |
- |
- |
- |
| 12 |
Hb_005714_050 |
0.242482863 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_1461s00200g [Populus trichocarpa] |
| 13 |
Hb_186158_010 |
0.2547430794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000175_580 |
0.2619419047 |
transcription factor, desease resistance |
TF Family: C3H, Gene Name: zf-CCCH |
hypothetical protein POPTR_0013s00890g [Populus trichocarpa] |
| 15 |
Hb_000414_150 |
0.2685606348 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 16 |
Hb_004241_010 |
0.2712219544 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 17 |
Hb_032974_010 |
0.2733568051 |
- |
- |
- |
| 18 |
Hb_002099_010 |
0.2803240061 |
- |
- |
cysteine proteinase inhibitor 5-like [Cucumis sativus] |
| 19 |
Hb_117092_010 |
0.2850154357 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 20 |
Hb_008215_030 |
0.2882605941 |
- |
- |
hypothetical protein 3 [Hevea brasiliensis] |