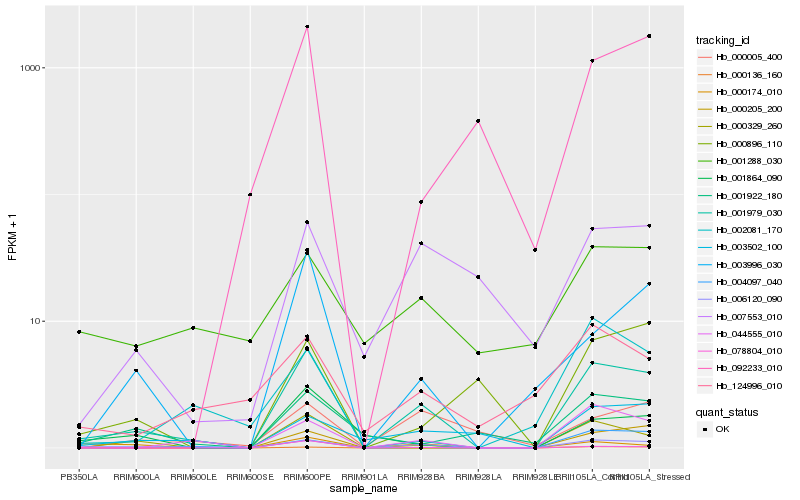
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001979_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s06130g [Populus trichocarpa] |
| 2 |
Hb_006120_090 |
0.1342395369 |
- |
- |
PREDICTED: desiccation-related protein PCC13-62-like [Jatropha curcas] |
| 3 |
Hb_000205_200 |
0.2365093 |
- |
- |
- |
| 4 |
Hb_044555_010 |
0.2407531341 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 5 |
Hb_003996_030 |
0.2765173719 |
- |
- |
- |
| 6 |
Hb_000329_260 |
0.2767474873 |
- |
- |
hypothetical protein MIMGU_mgv1a016162mg [Erythranthe guttata] |
| 7 |
Hb_001864_090 |
0.2885198317 |
- |
- |
- |
| 8 |
Hb_003502_100 |
0.293593385 |
- |
- |
- |
| 9 |
Hb_001922_180 |
0.2996072261 |
- |
- |
hevamine-A precursor, putative [Ricinus communis] |
| 10 |
Hb_007553_010 |
0.3012283266 |
- |
- |
- |
| 11 |
Hb_000174_010 |
0.3012955228 |
- |
- |
PREDICTED: uncharacterized protein LOC103444904 [Malus domestica] |
| 12 |
Hb_000005_400 |
0.3055359335 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 23 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000136_160 |
0.3090188468 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 14 |
Hb_000896_110 |
0.3090511235 |
- |
- |
- |
| 15 |
Hb_078804_010 |
0.3100734681 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_124996_010 |
0.3102279952 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 17 |
Hb_002081_170 |
0.3153286753 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 18 |
Hb_092233_010 |
0.3191303736 |
- |
- |
RecName: Full=Profilin-3; AltName: Full=Pollen allergen Hev b 8.0201; AltName: Allergen=Hev b 8.0201 [Hevea brasiliensis] |
| 19 |
Hb_001288_030 |
0.3198894185 |
- |
- |
- |
| 20 |
Hb_004097_040 |
0.3219257228 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |