| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
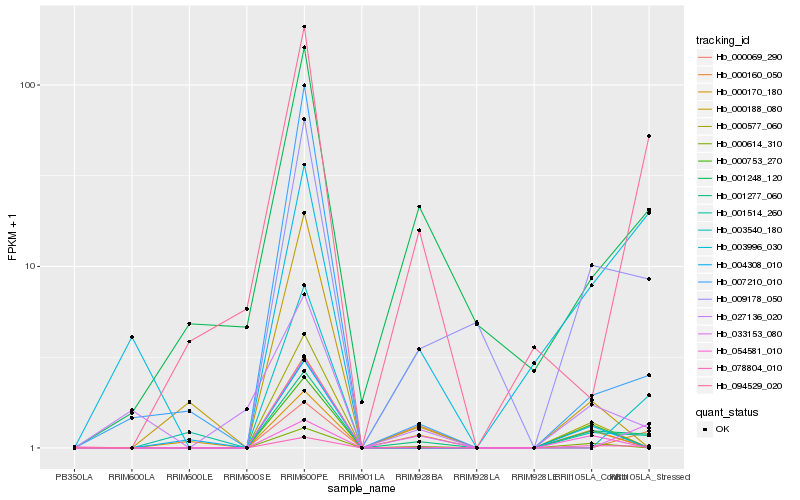
Hb_001277_060 |
0.0 |
- |
- |
ADP-ribosylation factor-like protein 5 [Morus notabilis] |
| 2 |
Hb_009178_050 |
0.1527008108 |
- |
- |
Elongation factor 1-alpha, putative [Ricinus communis] |
| 3 |
Hb_078804_010 |
0.1816559412 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_000614_310 |
0.2193355682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001248_120 |
0.2369399586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_033153_080 |
0.2483567129 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucoside 2''-O-glucosyltransferase-like [Populus euphratica] |
| 7 |
Hb_001514_260 |
0.2526091838 |
- |
- |
- |
| 8 |
Hb_000577_060 |
0.2583951412 |
- |
- |
- |
| 9 |
Hb_000160_050 |
0.2667688098 |
- |
- |
hypothetical protein JCGZ_02516 [Jatropha curcas] |
| 10 |
Hb_000753_270 |
0.2668402779 |
- |
- |
- |
| 11 |
Hb_094529_020 |
0.2788124147 |
- |
- |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 12 |
Hb_000069_290 |
0.281875463 |
- |
- |
hypothetical protein POPTR_0014s09500g [Populus trichocarpa] |
| 13 |
Hb_003540_180 |
0.2825087325 |
- |
- |
- |
| 14 |
Hb_007210_010 |
0.2885334356 |
- |
- |
- |
| 15 |
Hb_000188_080 |
0.2900470111 |
- |
- |
PREDICTED: uncharacterized protein LOC105629484 [Jatropha curcas] |
| 16 |
Hb_027136_020 |
0.290566452 |
- |
- |
- |
| 17 |
Hb_004308_010 |
0.2943353601 |
- |
- |
hypothetical protein CISIN_1g0004672mg, partial [Citrus sinensis] |
| 18 |
Hb_000170_180 |
0.2990454263 |
- |
- |
PREDICTED: uncharacterized protein LOC105109433 [Populus euphratica] |
| 19 |
Hb_003996_030 |
0.3004690285 |
- |
- |
- |
| 20 |
Hb_054581_010 |
0.3032898902 |
- |
- |
Type I inositol-1,4,5-trisphosphate 5-phosphatase CVP2 [Theobroma cacao] |