| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
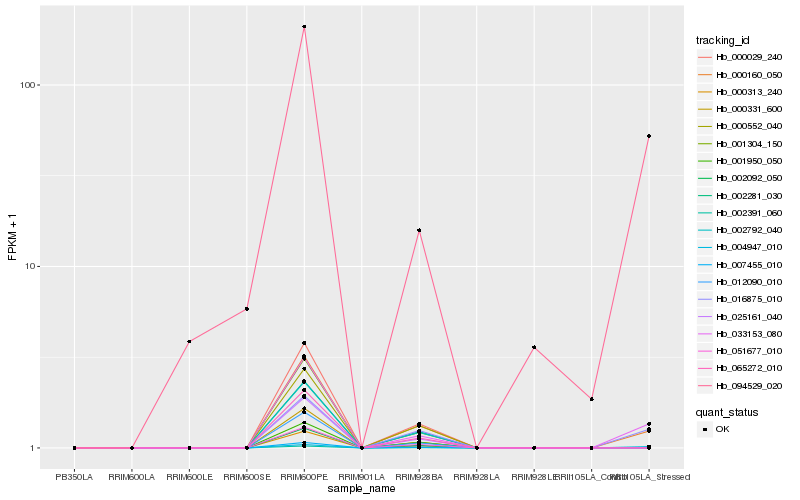
Hb_000160_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_02516 [Jatropha curcas] |
| 2 |
Hb_033153_080 |
0.0997432488 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucoside 2''-O-glucosyltransferase-like [Populus euphratica] |
| 3 |
Hb_016875_010 |
0.148581092 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 4 |
Hb_004947_010 |
0.1948125118 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 5 |
Hb_094529_020 |
0.2048722377 |
- |
- |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 6 |
Hb_051677_010 |
0.2110588539 |
- |
- |
Uncharacterized protein TCM_017953 [Theobroma cacao] |
| 7 |
Hb_002391_060 |
0.2110651844 |
- |
- |
PREDICTED: uncharacterized protein LOC100853799 [Vitis vinifera] |
| 8 |
Hb_007455_010 |
0.2110666717 |
- |
- |
PREDICTED: uncharacterized protein LOC105800996 [Gossypium raimondii] |
| 9 |
Hb_025161_040 |
0.2114740345 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_002792_040 |
0.2117294548 |
- |
- |
- |
| 11 |
Hb_000029_240 |
0.2119800947 |
- |
- |
PREDICTED: pre-mRNA-splicing factor SLU7-A-like [Elaeis guineensis] |
| 12 |
Hb_002281_030 |
0.2121156673 |
- |
- |
- |
| 13 |
Hb_065272_010 |
0.2125095772 |
- |
- |
hypothetical protein JCGZ_06349 [Jatropha curcas] |
| 14 |
Hb_002092_050 |
0.2130360672 |
- |
- |
hypothetical protein JCGZ_05877 [Jatropha curcas] |
| 15 |
Hb_000331_600 |
0.2134116838 |
- |
- |
PREDICTED: probable Ufm1-specific protease [Jatropha curcas] |
| 16 |
Hb_012090_010 |
0.2138385112 |
- |
- |
hypothetical protein RCOM_0904330 [Ricinus communis] |
| 17 |
Hb_000313_240 |
0.2139224721 |
- |
- |
hypothetical protein RCOM_0377590 [Ricinus communis] |
| 18 |
Hb_000552_040 |
0.2139458611 |
- |
- |
- |
| 19 |
Hb_001950_050 |
0.215112716 |
- |
- |
- |
| 20 |
Hb_001304_150 |
0.2161458139 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |