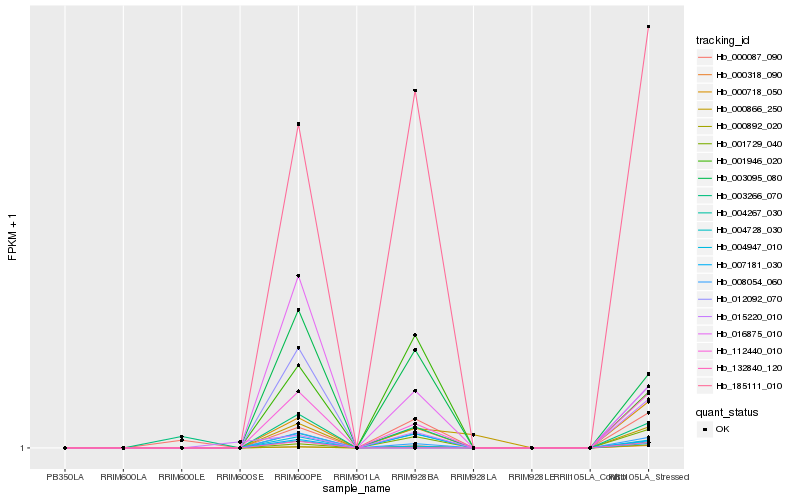
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_112440_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 2 |
Hb_008054_060 |
0.0722826506 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Populus euphratica] |
| 3 |
Hb_000718_050 |
0.1013948742 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X2 [Populus euphratica] |
| 4 |
Hb_185111_010 |
0.1714209874 |
- |
- |
- |
| 5 |
Hb_004947_010 |
0.1877460841 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 6 |
Hb_000892_020 |
0.1883116575 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_003095_080 |
0.1890288842 |
- |
- |
- |
| 8 |
Hb_016875_010 |
0.2189783022 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000318_090 |
0.2435560799 |
- |
- |
PREDICTED: uncharacterized protein LOC105179663 [Sesamum indicum] |
| 10 |
Hb_004267_030 |
0.2691976329 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_003266_070 |
0.2729202071 |
- |
- |
- |
| 12 |
Hb_000087_090 |
0.2819587393 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Jatropha curcas] |
| 13 |
Hb_007181_030 |
0.2823654377 |
- |
- |
PREDICTED: uncharacterized protein LOC105649502 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001946_020 |
0.2853268862 |
- |
- |
hypothetical protein RCOM_0224470 [Ricinus communis] |
| 15 |
Hb_004728_030 |
0.3024405984 |
- |
- |
hypothetical protein, partial [Lactobacillus brevis] |
| 16 |
Hb_132840_120 |
0.302850632 |
- |
- |
Vacuolar protein sorting-associated protein 8 like [Glycine soja] |
| 17 |
Hb_001729_040 |
0.3034195994 |
- |
- |
hypothetical protein glysoja_049826, partial [Glycine soja] |
| 18 |
Hb_015220_010 |
0.3154114854 |
- |
- |
PREDICTED: ribonuclease 3-like protein 3 [Jatropha curcas] |
| 19 |
Hb_012092_070 |
0.3188078803 |
- |
- |
PREDICTED: uncharacterized protein LOC105644084 [Jatropha curcas] |
| 20 |
Hb_000866_250 |
0.3240136878 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |