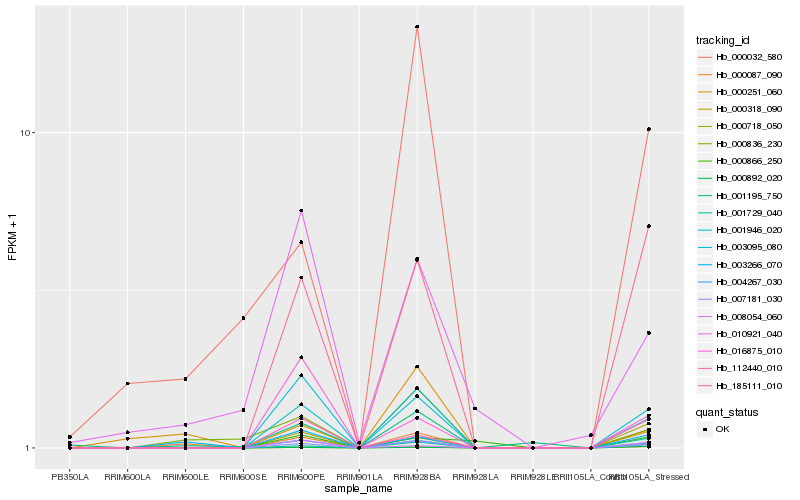
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007181_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649502 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001946_020 |
0.0415899234 |
- |
- |
hypothetical protein RCOM_0224470 [Ricinus communis] |
| 3 |
Hb_004267_030 |
0.1229179461 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_003095_080 |
0.172727174 |
- |
- |
- |
| 5 |
Hb_185111_010 |
0.1793938171 |
- |
- |
- |
| 6 |
Hb_000318_090 |
0.1931797431 |
- |
- |
PREDICTED: uncharacterized protein LOC105179663 [Sesamum indicum] |
| 7 |
Hb_000087_090 |
0.2595003258 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Jatropha curcas] |
| 8 |
Hb_000836_230 |
0.2683537523 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 9 |
Hb_000718_050 |
0.2694373281 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X2 [Populus euphratica] |
| 10 |
Hb_000032_580 |
0.2806569975 |
- |
- |
hypothetical protein RCOM_1311780 [Ricinus communis] |
| 11 |
Hb_112440_010 |
0.2823654377 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 12 |
Hb_001729_040 |
0.2829265816 |
- |
- |
hypothetical protein glysoja_049826, partial [Glycine soja] |
| 13 |
Hb_000892_020 |
0.2913810021 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_010921_040 |
0.3135570637 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 15 |
Hb_003266_070 |
0.3143309574 |
- |
- |
- |
| 16 |
Hb_008054_060 |
0.3152451181 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Populus euphratica] |
| 17 |
Hb_016875_010 |
0.3298978784 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 18 |
Hb_001195_750 |
0.3319739507 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 [Jatropha curcas] |
| 19 |
Hb_000866_250 |
0.3321062361 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 20 |
Hb_000251_060 |
0.3397827601 |
- |
- |
PREDICTED: uncharacterized protein LOC105649757 isoform X2 [Jatropha curcas] |