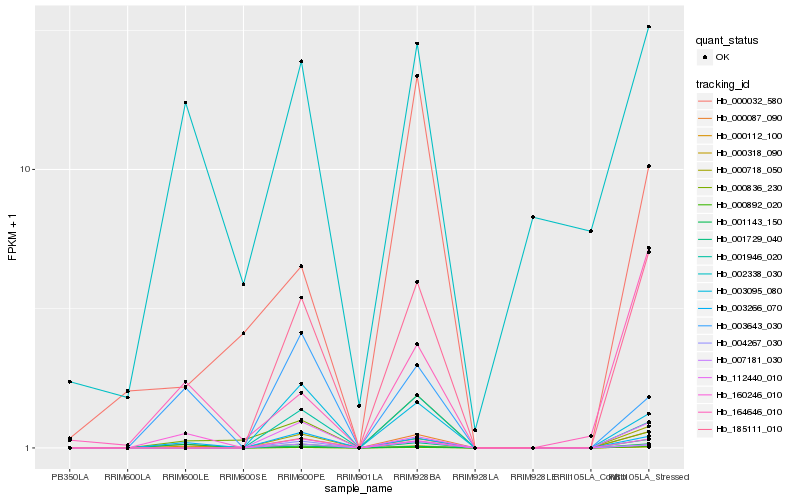
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000087_090 |
0.0 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Jatropha curcas] |
| 2 |
Hb_003266_070 |
0.1729637327 |
- |
- |
- |
| 3 |
Hb_185111_010 |
0.2053148727 |
- |
- |
- |
| 4 |
Hb_000318_090 |
0.2100460257 |
- |
- |
PREDICTED: uncharacterized protein LOC105179663 [Sesamum indicum] |
| 5 |
Hb_000892_020 |
0.2401287107 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000718_050 |
0.2423429435 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X2 [Populus euphratica] |
| 7 |
Hb_001729_040 |
0.248908985 |
- |
- |
hypothetical protein glysoja_049826, partial [Glycine soja] |
| 8 |
Hb_007181_030 |
0.2595003258 |
- |
- |
PREDICTED: uncharacterized protein LOC105649502 isoform X2 [Jatropha curcas] |
| 9 |
Hb_164646_010 |
0.2651145515 |
- |
- |
PREDICTED: acidic endochitinase-like [Pyrus x bretschneideri] |
| 10 |
Hb_001143_150 |
0.2774716081 |
- |
- |
PREDICTED: mitochondrial protein import protein MAS5-like [Populus euphratica] |
| 11 |
Hb_001946_020 |
0.2792188957 |
- |
- |
hypothetical protein RCOM_0224470 [Ricinus communis] |
| 12 |
Hb_112440_010 |
0.2819587393 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 13 |
Hb_003643_030 |
0.2840958479 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Populus euphratica] |
| 14 |
Hb_002338_030 |
0.2904309011 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P1-like [Tarenaya hassleriana] |
| 15 |
Hb_003095_080 |
0.2998297753 |
- |
- |
- |
| 16 |
Hb_000836_230 |
0.309537386 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 17 |
Hb_004267_030 |
0.3125408534 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_000032_580 |
0.3178068012 |
- |
- |
hypothetical protein RCOM_1311780 [Ricinus communis] |
| 19 |
Hb_160246_010 |
0.3219000436 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_000112_100 |
0.3236585749 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |