| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
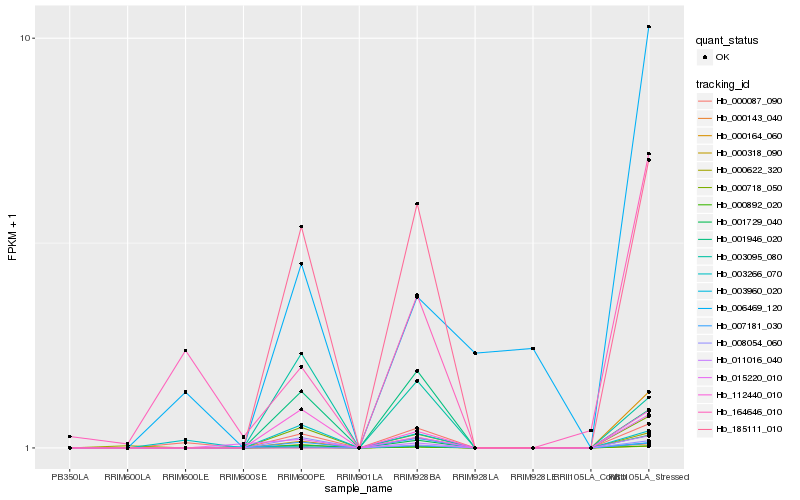
Hb_000892_020 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000718_050 |
0.087791613 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X2 [Populus euphratica] |
| 3 |
Hb_185111_010 |
0.1151543888 |
- |
- |
- |
| 4 |
Hb_000318_090 |
0.128925768 |
- |
- |
PREDICTED: uncharacterized protein LOC105179663 [Sesamum indicum] |
| 5 |
Hb_001729_040 |
0.1358604655 |
- |
- |
hypothetical protein glysoja_049826, partial [Glycine soja] |
| 6 |
Hb_112440_010 |
0.1883116575 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 7 |
Hb_015220_010 |
0.2047628501 |
- |
- |
PREDICTED: ribonuclease 3-like protein 3 [Jatropha curcas] |
| 8 |
Hb_000164_060 |
0.2149551061 |
- |
- |
- |
| 9 |
Hb_000087_090 |
0.2401287107 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Jatropha curcas] |
| 10 |
Hb_008054_060 |
0.2588478315 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Populus euphratica] |
| 11 |
Hb_003960_020 |
0.2773593392 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_007181_030 |
0.2913810021 |
- |
- |
PREDICTED: uncharacterized protein LOC105649502 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000622_320 |
0.2981391274 |
- |
- |
PREDICTED: chitinase 1-like [Vitis vinifera] |
| 14 |
Hb_164646_010 |
0.3061602678 |
- |
- |
PREDICTED: acidic endochitinase-like [Pyrus x bretschneideri] |
| 15 |
Hb_003095_080 |
0.3073791707 |
- |
- |
- |
| 16 |
Hb_006469_120 |
0.3080089501 |
- |
- |
hypothetical protein POPTR_0005s16430g [Populus trichocarpa] |
| 17 |
Hb_001946_020 |
0.3180025607 |
- |
- |
hypothetical protein RCOM_0224470 [Ricinus communis] |
| 18 |
Hb_003266_070 |
0.3357987967 |
- |
- |
- |
| 19 |
Hb_011016_040 |
0.344282291 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000143_040 |
0.3442823611 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |