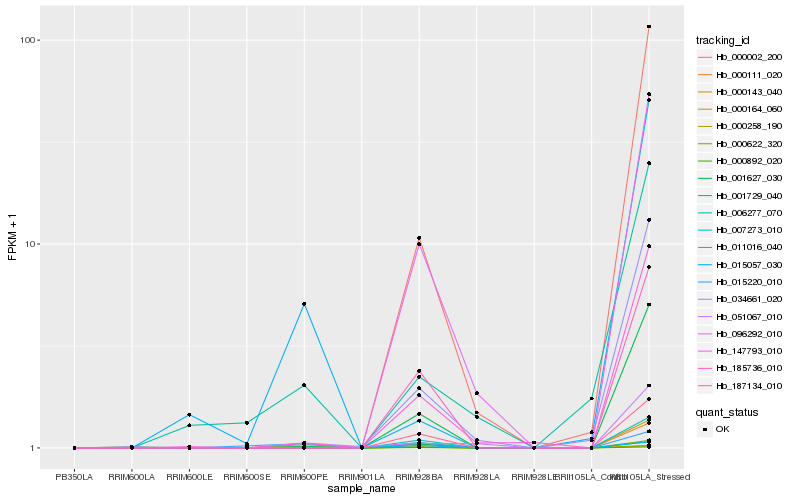
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000164_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_147793_010 |
0.2043749842 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PUB24-like [Jatropha curcas] |
| 3 |
Hb_000111_020 |
0.2066100215 |
- |
- |
hypothetical protein SORBIDRAFT_04g003755 [Sorghum bicolor] |
| 4 |
Hb_185736_010 |
0.2083296041 |
- |
- |
- |
| 5 |
Hb_001627_030 |
0.2104207597 |
- |
- |
hypothetical protein JCGZ_11752 [Jatropha curcas] |
| 6 |
Hb_007273_010 |
0.2105318413 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 7 |
Hb_187134_010 |
0.2124149402 |
- |
- |
- |
| 8 |
Hb_000892_020 |
0.2149551061 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_096292_010 |
0.2204155129 |
- |
- |
hypothetical protein JCGZ_25718 [Jatropha curcas] |
| 10 |
Hb_051067_010 |
0.2255427709 |
- |
- |
- |
| 11 |
Hb_001729_040 |
0.2261060303 |
- |
- |
hypothetical protein glysoja_049826, partial [Glycine soja] |
| 12 |
Hb_000002_200 |
0.2276785333 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF096 [Brassica rapa] |
| 13 |
Hb_015220_010 |
0.2322217096 |
- |
- |
PREDICTED: ribonuclease 3-like protein 3 [Jatropha curcas] |
| 14 |
Hb_006277_070 |
0.2343351587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_015057_030 |
0.2399193727 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-52, partial [Jatropha curcas] |
| 16 |
Hb_034661_020 |
0.2404000662 |
- |
- |
hypothetical protein JCGZ_04352 [Jatropha curcas] |
| 17 |
Hb_000622_320 |
0.2526898839 |
- |
- |
PREDICTED: chitinase 1-like [Vitis vinifera] |
| 18 |
Hb_011016_040 |
0.2529235826 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 19 |
Hb_000143_040 |
0.2529374064 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 20 |
Hb_000258_190 |
0.2532570758 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |