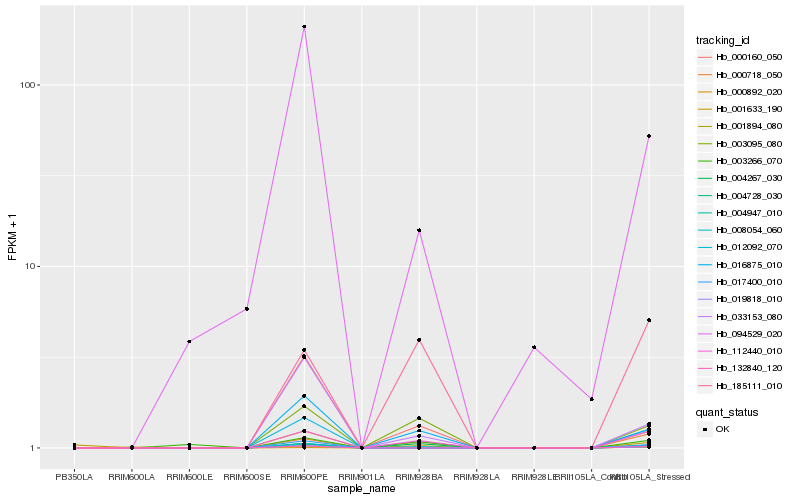
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008054_060 |
0.0 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Populus euphratica] |
| 2 |
Hb_112440_010 |
0.0722826506 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 3 |
Hb_004947_010 |
0.1161128953 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 4 |
Hb_016875_010 |
0.1613937647 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000718_050 |
0.1730445987 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X2 [Populus euphratica] |
| 6 |
Hb_003095_080 |
0.1850304076 |
- |
- |
- |
| 7 |
Hb_185111_010 |
0.2355449028 |
- |
- |
- |
| 8 |
Hb_000892_020 |
0.2588478315 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_094529_020 |
0.2644287893 |
- |
- |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 10 |
Hb_033153_080 |
0.2682608795 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucoside 2''-O-glucosyltransferase-like [Populus euphratica] |
| 11 |
Hb_004267_030 |
0.2740303913 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_132840_120 |
0.2752729638 |
- |
- |
Vacuolar protein sorting-associated protein 8 like [Glycine soja] |
| 13 |
Hb_012092_070 |
0.2756410216 |
- |
- |
PREDICTED: uncharacterized protein LOC105644084 [Jatropha curcas] |
| 14 |
Hb_003266_070 |
0.2809206041 |
- |
- |
- |
| 15 |
Hb_019818_010 |
0.2824642497 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 16 |
Hb_001633_190 |
0.2860499475 |
- |
- |
PREDICTED: probable methyltransferase PMT11 [Jatropha curcas] |
| 17 |
Hb_004728_030 |
0.2885201124 |
- |
- |
hypothetical protein, partial [Lactobacillus brevis] |
| 18 |
Hb_017400_010 |
0.2926626067 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_000160_050 |
0.2931394401 |
- |
- |
hypothetical protein JCGZ_02516 [Jatropha curcas] |
| 20 |
Hb_001894_080 |
0.3075599929 |
- |
- |
ATP binding protein, putative [Ricinus communis] |