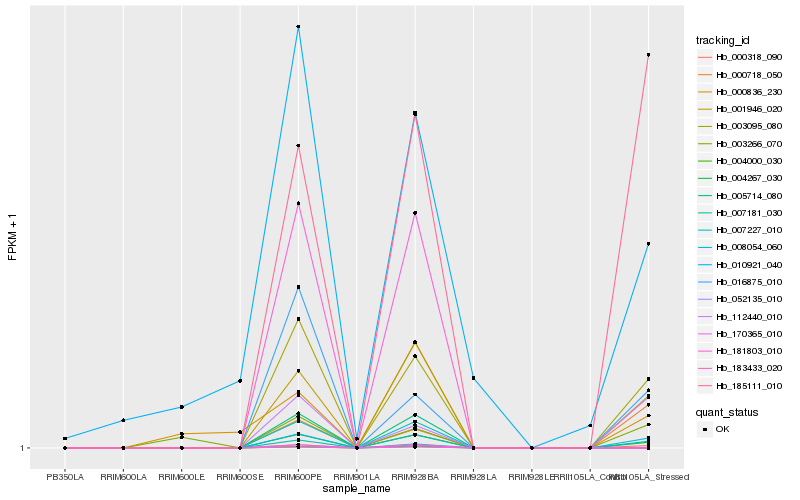
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004267_030 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_001946_020 |
0.0847173125 |
- |
- |
hypothetical protein RCOM_0224470 [Ricinus communis] |
| 3 |
Hb_003095_080 |
0.0923690154 |
- |
- |
- |
| 4 |
Hb_007181_030 |
0.1229179461 |
- |
- |
PREDICTED: uncharacterized protein LOC105649502 isoform X2 [Jatropha curcas] |
| 5 |
Hb_016875_010 |
0.2339616546 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_185111_010 |
0.243211853 |
- |
- |
- |
| 7 |
Hb_010921_040 |
0.2559575192 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 8 |
Hb_112440_010 |
0.2691976329 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 9 |
Hb_008054_060 |
0.2740303913 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Populus euphratica] |
| 10 |
Hb_000318_090 |
0.2853989054 |
- |
- |
PREDICTED: uncharacterized protein LOC105179663 [Sesamum indicum] |
| 11 |
Hb_000836_230 |
0.2866880844 |
- |
- |
Uncharacterized protein TCM_041879 [Theobroma cacao] |
| 12 |
Hb_000718_050 |
0.2992111267 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X2 [Populus euphratica] |
| 13 |
Hb_003266_070 |
0.2995193093 |
- |
- |
- |
| 14 |
Hb_007227_010 |
0.3022204245 |
- |
- |
hypothetical protein BN927_00890 [Lactococcus lactis subsp. lactis Dephy 1] |
| 15 |
Hb_005714_080 |
0.3022495005 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC105109431 [Populus euphratica] |
| 16 |
Hb_052135_010 |
0.3022776882 |
- |
- |
PREDICTED: phytochrome B isoform X2 [Jatropha curcas] |
| 17 |
Hb_170365_010 |
0.302279555 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 18 |
Hb_183433_020 |
0.3022914202 |
- |
- |
PREDICTED: uncharacterized protein LOC105851424 [Cicer arietinum] |
| 19 |
Hb_181803_010 |
0.3023185238 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 20 |
Hb_004000_030 |
0.3023203219 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |