| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
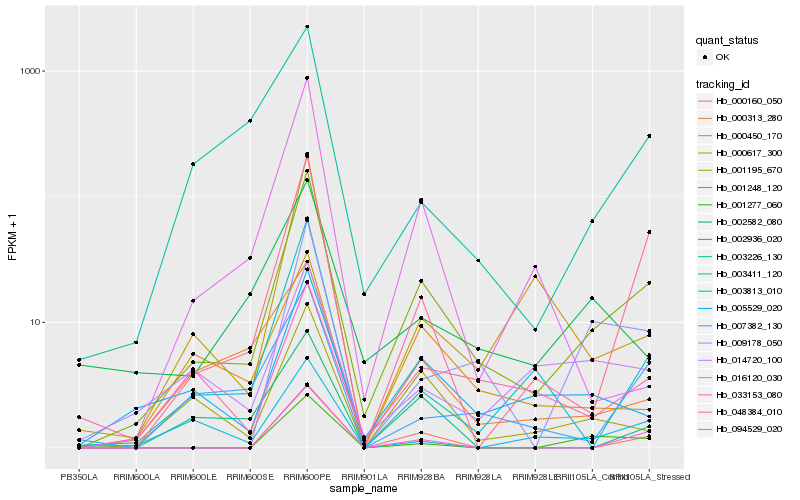
Hb_001248_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_094529_020 |
0.1976024595 |
- |
- |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 3 |
Hb_005529_020 |
0.2114957332 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 4 |
Hb_001195_670 |
0.2130892592 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |
| 5 |
Hb_000160_050 |
0.2275290671 |
- |
- |
hypothetical protein JCGZ_02516 [Jatropha curcas] |
| 6 |
Hb_003411_120 |
0.2310588749 |
- |
- |
hypothetical protein POPTR_0010s13670g [Populus trichocarpa] |
| 7 |
Hb_000450_170 |
0.231602569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_009178_050 |
0.2332273192 |
- |
- |
Elongation factor 1-alpha, putative [Ricinus communis] |
| 9 |
Hb_001277_060 |
0.2369399586 |
- |
- |
ADP-ribosylation factor-like protein 5 [Morus notabilis] |
| 10 |
Hb_007382_130 |
0.2371377619 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 11 |
Hb_033153_080 |
0.2373702473 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucoside 2''-O-glucosyltransferase-like [Populus euphratica] |
| 12 |
Hb_000313_280 |
0.2378629343 |
- |
- |
hypothetical protein PRUPE_ppa014856mg, partial [Prunus persica] |
| 13 |
Hb_003226_130 |
0.2402971429 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 14 |
Hb_000617_300 |
0.2415893159 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 15 |
Hb_014720_100 |
0.242761508 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 16 |
Hb_002582_080 |
0.2484692698 |
- |
- |
- |
| 17 |
Hb_002936_020 |
0.2544597944 |
- |
- |
hypothetical protein VITISV_001748 [Vitis vinifera] |
| 18 |
Hb_048384_010 |
0.2558160593 |
- |
- |
PREDICTED: probable receptor-like protein kinase At2g42960 [Cucumis sativus] |
| 19 |
Hb_003813_010 |
0.2576927877 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 1-like [Jatropha curcas] |
| 20 |
Hb_016120_030 |
0.2578764323 |
- |
- |
hypothetical protein EUGRSUZ_D00080 [Eucalyptus grandis] |