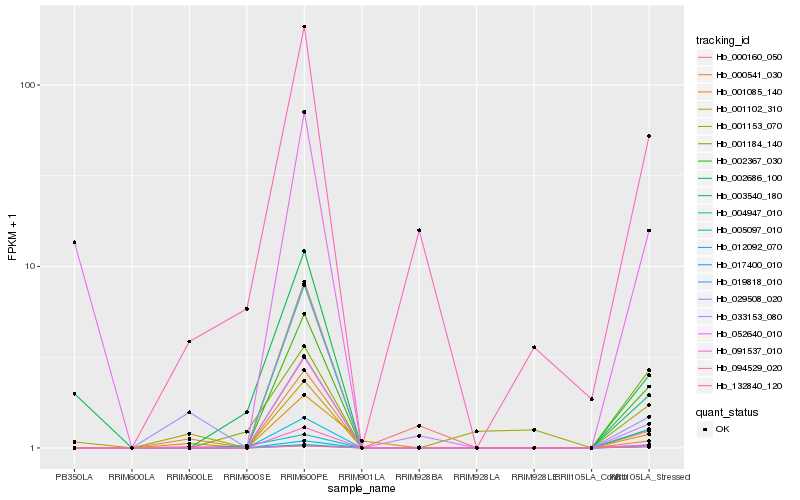
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002367_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s30725g [Populus trichocarpa] |
| 2 |
Hb_017400_010 |
0.0770597531 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_003540_180 |
0.0994155113 |
- |
- |
- |
| 4 |
Hb_019818_010 |
0.1111748214 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 5 |
Hb_001102_310 |
0.142426502 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 6 |
Hb_012092_070 |
0.1465345036 |
- |
- |
PREDICTED: uncharacterized protein LOC105644084 [Jatropha curcas] |
| 7 |
Hb_091537_010 |
0.1846397098 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_033153_080 |
0.1925063266 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucoside 2''-O-glucosyltransferase-like [Populus euphratica] |
| 9 |
Hb_001085_140 |
0.2016925298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_094529_020 |
0.2167708991 |
- |
- |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 11 |
Hb_132840_120 |
0.2225810861 |
- |
- |
Vacuolar protein sorting-associated protein 8 like [Glycine soja] |
| 12 |
Hb_001184_140 |
0.2316383335 |
- |
- |
hypothetical protein B456_008G022200 [Gossypium raimondii] |
| 13 |
Hb_004947_010 |
0.2413614669 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 14 |
Hb_001153_070 |
0.2534875688 |
- |
- |
- |
| 15 |
Hb_002686_100 |
0.2585437255 |
- |
- |
- |
| 16 |
Hb_029508_020 |
0.2604476023 |
- |
- |
- |
| 17 |
Hb_052640_010 |
0.2627774421 |
- |
- |
PREDICTED: putative quinone-oxidoreductase homolog, chloroplastic [Jatropha curcas] |
| 18 |
Hb_005097_010 |
0.2666579033 |
- |
- |
PREDICTED: aluminum-activated malate transporter 7-like isoform X1 [Populus euphratica] |
| 19 |
Hb_000541_030 |
0.2679250486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000160_050 |
0.2783316382 |
- |
- |
hypothetical protein JCGZ_02516 [Jatropha curcas] |