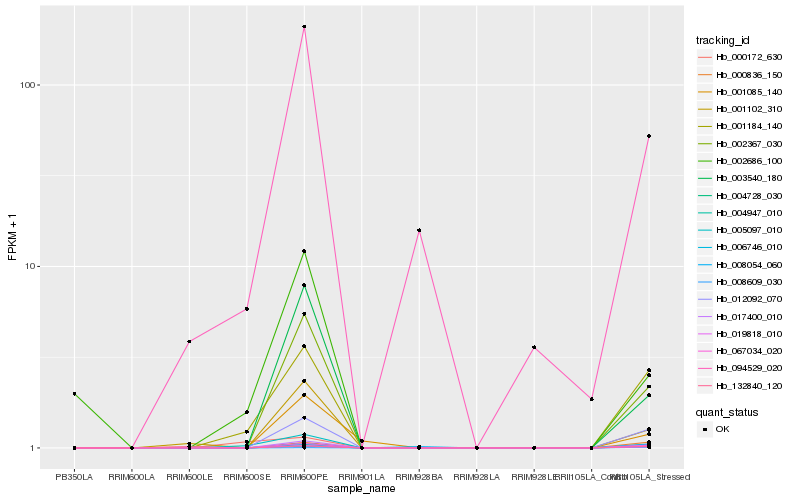
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001184_140 |
0.0 |
- |
- |
hypothetical protein B456_008G022200 [Gossypium raimondii] |
| 2 |
Hb_012092_070 |
0.1620857443 |
- |
- |
PREDICTED: uncharacterized protein LOC105644084 [Jatropha curcas] |
| 3 |
Hb_132840_120 |
0.169240125 |
- |
- |
Vacuolar protein sorting-associated protein 8 like [Glycine soja] |
| 4 |
Hb_019818_010 |
0.1705628324 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 5 |
Hb_017400_010 |
0.1847332142 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_004728_030 |
0.1962571782 |
- |
- |
hypothetical protein, partial [Lactobacillus brevis] |
| 7 |
Hb_002367_030 |
0.2316383335 |
- |
- |
hypothetical protein POPTR_0001s30725g [Populus trichocarpa] |
| 8 |
Hb_094529_020 |
0.2713355706 |
- |
- |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 9 |
Hb_005097_010 |
0.2752823564 |
- |
- |
PREDICTED: aluminum-activated malate transporter 7-like isoform X1 [Populus euphratica] |
| 10 |
Hb_006746_010 |
0.2819986422 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 11 |
Hb_004947_010 |
0.291159655 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 12 |
Hb_000836_150 |
0.294809729 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 13 |
Hb_001102_310 |
0.2970564925 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 14 |
Hb_067034_020 |
0.303464947 |
- |
- |
PREDICTED: iridoid synthase [Jatropha curcas] |
| 15 |
Hb_003540_180 |
0.3078113474 |
- |
- |
- |
| 16 |
Hb_000172_630 |
0.3080256857 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 17 |
Hb_008054_060 |
0.3127659221 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Populus euphratica] |
| 18 |
Hb_002686_100 |
0.318704931 |
- |
- |
- |
| 19 |
Hb_001085_140 |
0.3257088615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_008609_030 |
0.3258914934 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |