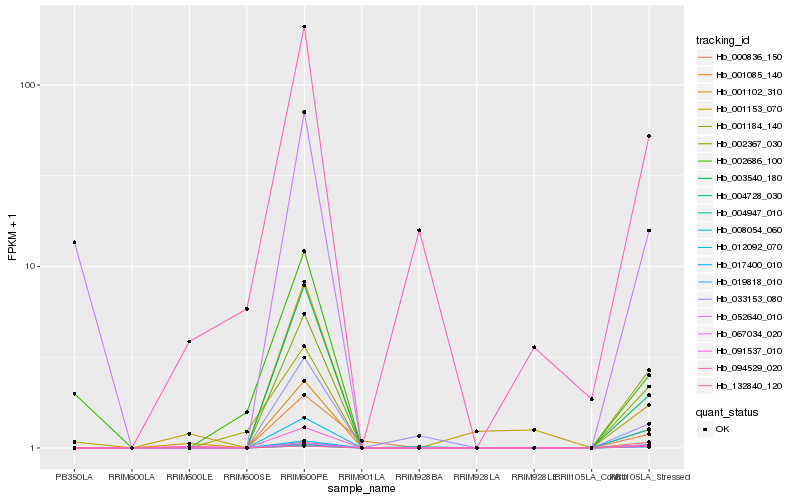
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017400_010 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_019818_010 |
0.0342708626 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 3 |
Hb_012092_070 |
0.069874356 |
- |
- |
PREDICTED: uncharacterized protein LOC105644084 [Jatropha curcas] |
| 4 |
Hb_002367_030 |
0.0770597531 |
- |
- |
hypothetical protein POPTR_0001s30725g [Populus trichocarpa] |
| 5 |
Hb_132840_120 |
0.1466833803 |
- |
- |
Vacuolar protein sorting-associated protein 8 like [Glycine soja] |
| 6 |
Hb_003540_180 |
0.1754918514 |
- |
- |
- |
| 7 |
Hb_001102_310 |
0.1829846862 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 8 |
Hb_001184_140 |
0.1847332142 |
- |
- |
hypothetical protein B456_008G022200 [Gossypium raimondii] |
| 9 |
Hb_004728_030 |
0.206225388 |
- |
- |
hypothetical protein, partial [Lactobacillus brevis] |
| 10 |
Hb_004947_010 |
0.2298019668 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 11 |
Hb_001085_140 |
0.2306110812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_033153_080 |
0.2311981924 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucoside 2''-O-glucosyltransferase-like [Populus euphratica] |
| 13 |
Hb_094529_020 |
0.2329721651 |
- |
- |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 14 |
Hb_091537_010 |
0.2334175483 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_067034_020 |
0.2689401874 |
- |
- |
PREDICTED: iridoid synthase [Jatropha curcas] |
| 16 |
Hb_052640_010 |
0.2822957906 |
- |
- |
PREDICTED: putative quinone-oxidoreductase homolog, chloroplastic [Jatropha curcas] |
| 17 |
Hb_008054_060 |
0.2926626067 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Populus euphratica] |
| 18 |
Hb_002686_100 |
0.2945099715 |
- |
- |
- |
| 19 |
Hb_001153_070 |
0.2995284857 |
- |
- |
- |
| 20 |
Hb_000836_150 |
0.2998077987 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |