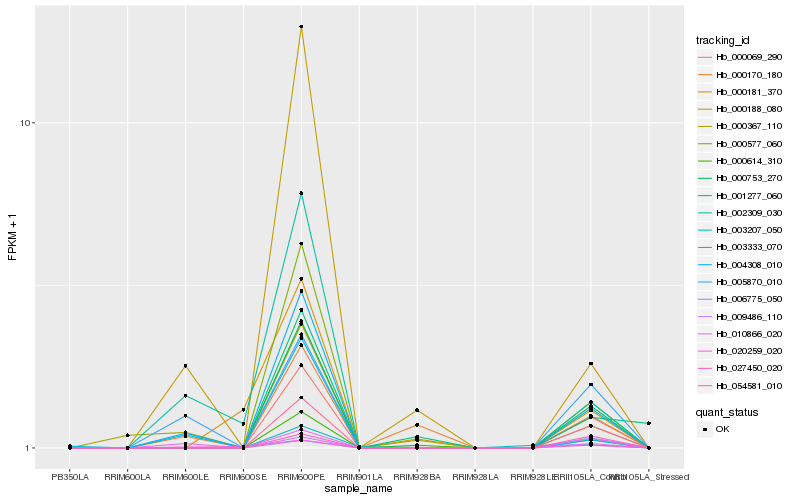
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_290 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s09500g [Populus trichocarpa] |
| 2 |
Hb_000753_270 |
0.0458844409 |
- |
- |
- |
| 3 |
Hb_054581_010 |
0.0476759147 |
- |
- |
Type I inositol-1,4,5-trisphosphate 5-phosphatase CVP2 [Theobroma cacao] |
| 4 |
Hb_000577_060 |
0.1488601716 |
- |
- |
- |
| 5 |
Hb_000614_310 |
0.1756065869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004308_010 |
0.1850901795 |
- |
- |
hypothetical protein CISIN_1g0004672mg, partial [Citrus sinensis] |
| 7 |
Hb_027450_020 |
0.197940009 |
- |
- |
PREDICTED: B3 domain-containing protein At2g31420-like [Citrus sinensis] |
| 8 |
Hb_009486_110 |
0.1996969909 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 9 |
Hb_005870_010 |
0.2652697554 |
- |
- |
- |
| 10 |
Hb_001277_060 |
0.281875463 |
- |
- |
ADP-ribosylation factor-like protein 5 [Morus notabilis] |
| 11 |
Hb_000181_370 |
0.2878452856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006775_050 |
0.2935946675 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g29670 isoform X2 [Nicotiana sylvestris] |
| 13 |
Hb_010866_020 |
0.294246785 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000188_080 |
0.2950717934 |
- |
- |
PREDICTED: uncharacterized protein LOC105629484 [Jatropha curcas] |
| 15 |
Hb_020259_020 |
0.2966963078 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 4 [Jatropha curcas] |
| 16 |
Hb_003207_050 |
0.2991286367 |
- |
- |
PREDICTED: cucumisin-like [Jatropha curcas] |
| 17 |
Hb_000170_180 |
0.3000261312 |
- |
- |
PREDICTED: uncharacterized protein LOC105109433 [Populus euphratica] |
| 18 |
Hb_002309_030 |
0.3028029783 |
- |
- |
Glycerol dehydrogenase large subunit, putative [Theobroma cacao] |
| 19 |
Hb_000367_110 |
0.3094292077 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic isoform X1 [Sesamum indicum] |
| 20 |
Hb_003333_070 |
0.3100221708 |
- |
- |
Ent-kaurenoic acid oxidase, putative [Ricinus communis] |