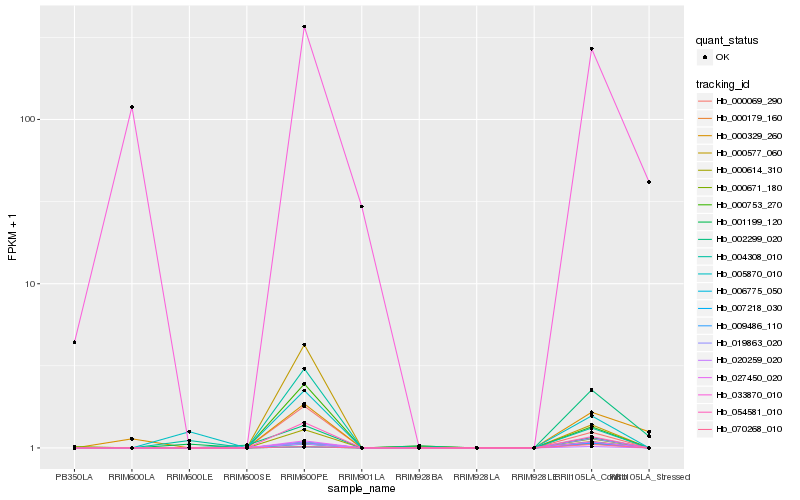
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027450_020 |
0.0 |
- |
- |
PREDICTED: B3 domain-containing protein At2g31420-like [Citrus sinensis] |
| 2 |
Hb_009486_110 |
0.0017843968 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 3 |
Hb_006775_050 |
0.0977354603 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g29670 isoform X2 [Nicotiana sylvestris] |
| 4 |
Hb_019863_020 |
0.1496200537 |
- |
- |
PREDICTED: wound-induced protein 1 [Jatropha curcas] |
| 5 |
Hb_054581_010 |
0.1508993573 |
- |
- |
Type I inositol-1,4,5-trisphosphate 5-phosphatase CVP2 [Theobroma cacao] |
| 6 |
Hb_000069_290 |
0.197940009 |
- |
- |
hypothetical protein POPTR_0014s09500g [Populus trichocarpa] |
| 7 |
Hb_000753_270 |
0.24272966 |
- |
- |
- |
| 8 |
Hb_070268_010 |
0.2509984976 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 9 |
Hb_005870_010 |
0.2817722398 |
- |
- |
- |
| 10 |
Hb_000614_310 |
0.3066601324 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000329_260 |
0.3349952118 |
- |
- |
hypothetical protein MIMGU_mgv1a016162mg [Erythranthe guttata] |
| 12 |
Hb_004308_010 |
0.3353320931 |
- |
- |
hypothetical protein CISIN_1g0004672mg, partial [Citrus sinensis] |
| 13 |
Hb_020259_020 |
0.3403971017 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 4 [Jatropha curcas] |
| 14 |
Hb_000577_060 |
0.3410884403 |
- |
- |
- |
| 15 |
Hb_000179_160 |
0.3515830509 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-3-like [Jatropha curcas] |
| 16 |
Hb_001199_120 |
0.3541760911 |
- |
- |
PREDICTED: uncharacterized protein LOC103437171 [Malus domestica] |
| 17 |
Hb_033870_010 |
0.3564429343 |
- |
- |
PREDICTED: uncharacterized protein LOC104101109 isoform X1 [Nicotiana tomentosiformis] |
| 18 |
Hb_007218_030 |
0.3660464117 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Jatropha curcas] |
| 19 |
Hb_000671_180 |
0.3743142701 |
- |
- |
RING zinc finger protein, putative [Arabidopsis thaliana] |
| 20 |
Hb_002299_020 |
0.3743506828 |
- |
- |
conserved hypothetical protein [Ricinus communis] |