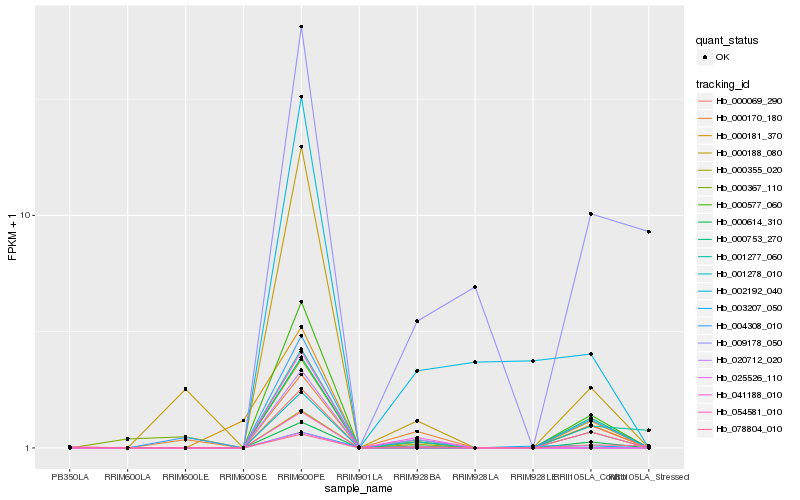
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000614_310 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000753_270 |
0.1638080392 |
- |
- |
- |
| 3 |
Hb_000069_290 |
0.1756065869 |
- |
- |
hypothetical protein POPTR_0014s09500g [Populus trichocarpa] |
| 4 |
Hb_000577_060 |
0.1813262151 |
- |
- |
- |
| 5 |
Hb_000170_180 |
0.1953624829 |
- |
- |
PREDICTED: uncharacterized protein LOC105109433 [Populus euphratica] |
| 6 |
Hb_054581_010 |
0.198189577 |
- |
- |
Type I inositol-1,4,5-trisphosphate 5-phosphatase CVP2 [Theobroma cacao] |
| 7 |
Hb_001277_060 |
0.2193355682 |
- |
- |
ADP-ribosylation factor-like protein 5 [Morus notabilis] |
| 8 |
Hb_004308_010 |
0.2231401515 |
- |
- |
hypothetical protein CISIN_1g0004672mg, partial [Citrus sinensis] |
| 9 |
Hb_000188_080 |
0.2433881199 |
- |
- |
PREDICTED: uncharacterized protein LOC105629484 [Jatropha curcas] |
| 10 |
Hb_000181_370 |
0.2485546561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000367_110 |
0.2572111742 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic isoform X1 [Sesamum indicum] |
| 12 |
Hb_002192_040 |
0.2595841214 |
- |
- |
- |
| 13 |
Hb_009178_050 |
0.2707036524 |
- |
- |
Elongation factor 1-alpha, putative [Ricinus communis] |
| 14 |
Hb_003207_050 |
0.2724000489 |
- |
- |
PREDICTED: cucumisin-like [Jatropha curcas] |
| 15 |
Hb_078804_010 |
0.2944040343 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_041188_010 |
0.2944618474 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Nicotiana tomentosiformis] |
| 17 |
Hb_025526_110 |
0.2945143019 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 18 |
Hb_020712_020 |
0.2948187949 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Elaeis guineensis] |
| 19 |
Hb_000355_020 |
0.2953927767 |
- |
- |
- |
| 20 |
Hb_001278_010 |
0.2955820733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |