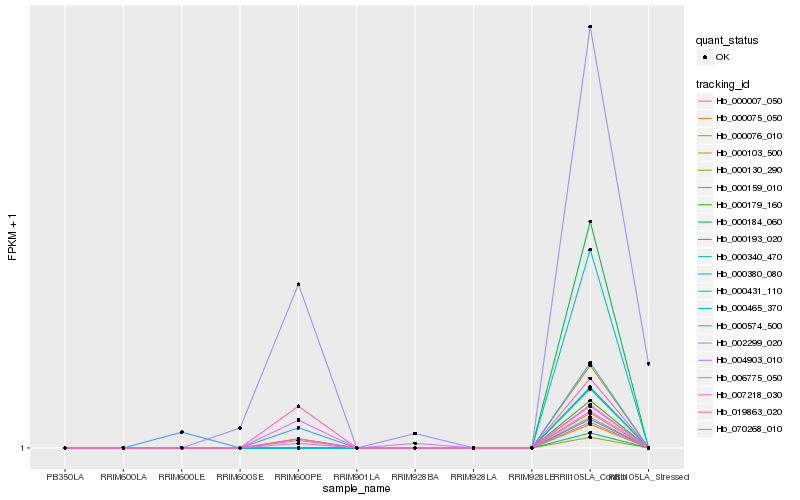
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000179_160 |
0.0 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-3-like [Jatropha curcas] |
| 2 |
Hb_007218_030 |
0.0155476105 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Jatropha curcas] |
| 3 |
Hb_070268_010 |
0.1048924346 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 4 |
Hb_019863_020 |
0.2065025594 |
- |
- |
PREDICTED: wound-induced protein 1 [Jatropha curcas] |
| 5 |
Hb_004903_010 |
0.2244057421 |
- |
- |
- |
| 6 |
Hb_000574_500 |
0.257017173 |
- |
- |
hypothetical protein JCGZ_06123 [Jatropha curcas] |
| 7 |
Hb_006775_050 |
0.2573787574 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g29670 isoform X2 [Nicotiana sylvestris] |
| 8 |
Hb_002299_020 |
0.272057992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000007_050 |
0.2885527553 |
- |
- |
- |
| 10 |
Hb_000075_050 |
0.2885527553 |
- |
- |
- |
| 11 |
Hb_000076_010 |
0.2885527553 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 12 |
Hb_000103_500 |
0.2885527553 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000130_290 |
0.2885527553 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000159_010 |
0.2885527553 |
- |
- |
- |
| 15 |
Hb_000184_060 |
0.2885527553 |
- |
- |
- |
| 16 |
Hb_000193_020 |
0.2885527553 |
- |
- |
- |
| 17 |
Hb_000340_470 |
0.2885527553 |
- |
- |
- |
| 18 |
Hb_000380_080 |
0.2885527553 |
- |
- |
hypothetical protein JCGZ_05489 [Jatropha curcas] |
| 19 |
Hb_000431_110 |
0.2885527553 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 20 |
Hb_000465_370 |
0.2885527553 |
- |
- |
hypothetical protein CISIN_1g039324mg, partial [Citrus sinensis] |