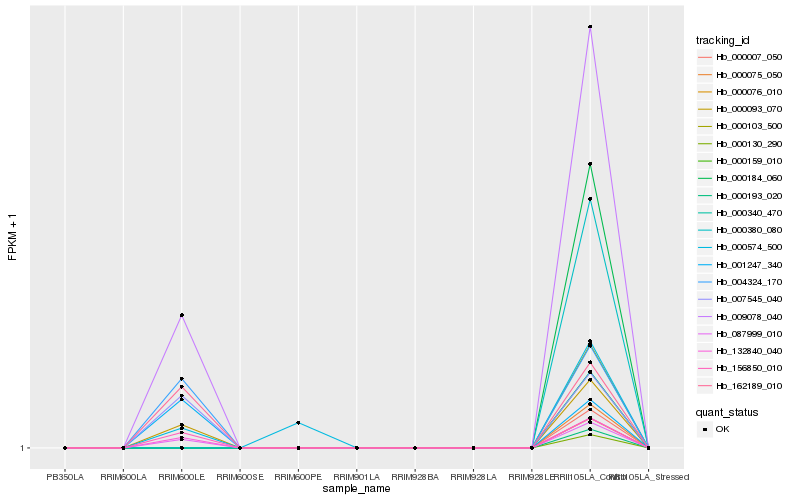
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009078_040 |
0.0 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 2 |
Hb_087999_010 |
0.0483931922 |
- |
- |
PREDICTED: uncharacterized protein LOC105640118 [Jatropha curcas] |
| 3 |
Hb_000093_070 |
0.0491022534 |
transcription factor |
TF Family: C2C2-YABBY |
Protein CRABS CLAW, putative [Ricinus communis] |
| 4 |
Hb_132840_040 |
0.0586406595 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase, putative [Ricinus communis] |
| 5 |
Hb_156850_010 |
0.1326142286 |
- |
- |
PREDICTED: uncharacterized protein LOC105635403 [Jatropha curcas] |
| 6 |
Hb_004324_170 |
0.1857541041 |
- |
- |
- |
| 7 |
Hb_007545_040 |
0.1928088965 |
- |
- |
PREDICTED: uncharacterized protein LOC104884077 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_162189_010 |
0.1986954884 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 9 |
Hb_001247_340 |
0.2719210184 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 10 |
Hb_000574_500 |
0.2958064967 |
- |
- |
hypothetical protein JCGZ_06123 [Jatropha curcas] |
| 11 |
Hb_000007_050 |
0.3277438774 |
- |
- |
- |
| 12 |
Hb_000075_050 |
0.3277438774 |
- |
- |
- |
| 13 |
Hb_000076_010 |
0.3277438774 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 14 |
Hb_000103_500 |
0.3277438774 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_000130_290 |
0.3277438774 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000159_010 |
0.3277438774 |
- |
- |
- |
| 17 |
Hb_000184_060 |
0.3277438774 |
- |
- |
- |
| 18 |
Hb_000193_020 |
0.3277438774 |
- |
- |
- |
| 19 |
Hb_000340_470 |
0.3277438774 |
- |
- |
- |
| 20 |
Hb_000380_080 |
0.3277438774 |
- |
- |
hypothetical protein JCGZ_05489 [Jatropha curcas] |