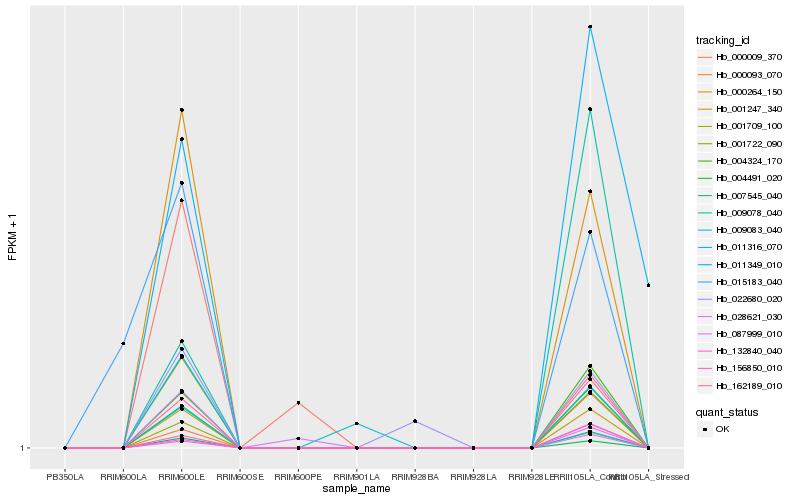
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001247_340 |
0.0 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 2 |
Hb_004491_020 |
0.0667425102 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 3 |
Hb_162189_010 |
0.0748460483 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 4 |
Hb_000264_150 |
0.0757615079 |
- |
- |
PREDICTED: uncharacterized protein LOC103870759 [Brassica rapa] |
| 5 |
Hb_007545_040 |
0.0808185596 |
- |
- |
PREDICTED: uncharacterized protein LOC104884077 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_004324_170 |
0.0879671876 |
- |
- |
- |
| 7 |
Hb_001722_090 |
0.1061347937 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 8 |
Hb_001709_100 |
0.106985837 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 9 |
Hb_156850_010 |
0.1414777482 |
- |
- |
PREDICTED: uncharacterized protein LOC105635403 [Jatropha curcas] |
| 10 |
Hb_132840_040 |
0.2148562272 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase, putative [Ricinus communis] |
| 11 |
Hb_000093_070 |
0.224210695 |
transcription factor |
TF Family: C2C2-YABBY |
Protein CRABS CLAW, putative [Ricinus communis] |
| 12 |
Hb_087999_010 |
0.2249050176 |
- |
- |
PREDICTED: uncharacterized protein LOC105640118 [Jatropha curcas] |
| 13 |
Hb_022680_020 |
0.2616157258 |
- |
- |
PREDICTED: MLP-like protein 423 [Jatropha curcas] |
| 14 |
Hb_009078_040 |
0.2719210184 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 15 |
Hb_015183_040 |
0.2946438048 |
- |
- |
- |
| 16 |
Hb_011349_010 |
0.2964441629 |
- |
- |
PREDICTED: MLP-like protein 423 [Jatropha curcas] |
| 17 |
Hb_009083_040 |
0.3288146411 |
- |
- |
hypothetical protein JCGZ_12563 [Jatropha curcas] |
| 18 |
Hb_011316_070 |
0.3319020671 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000009_370 |
0.3535052098 |
- |
- |
hypothetical protein JCGZ_12284 [Jatropha curcas] |
| 20 |
Hb_028621_030 |
0.3665240985 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |