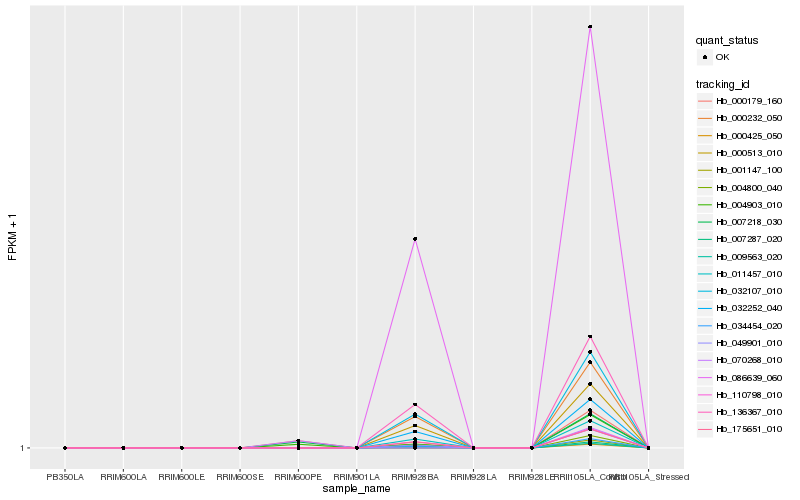
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004903_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_034454_020 |
0.219359859 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_049901_010 |
0.2195065602 |
- |
- |
PREDICTED: uncharacterized protein LOC105766879 [Gossypium raimondii] |
| 4 |
Hb_007218_030 |
0.2197585255 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Jatropha curcas] |
| 5 |
Hb_000179_160 |
0.2244057421 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-3-like [Jatropha curcas] |
| 6 |
Hb_000425_050 |
0.2670257397 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 7 |
Hb_009563_020 |
0.2670966138 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_007287_020 |
0.2672312074 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 9 |
Hb_001147_100 |
0.2674120237 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 10 |
Hb_004800_040 |
0.267837428 |
- |
- |
PREDICTED: probable polyol transporter 6 [Jatropha curcas] |
| 11 |
Hb_175651_010 |
0.2684591677 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 12 |
Hb_110798_010 |
0.2685043534 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 13 |
Hb_011457_010 |
0.2692989144 |
- |
- |
nucleolar protein nop56, putative [Ricinus communis] |
| 14 |
Hb_032252_040 |
0.2716229356 |
- |
- |
- |
| 15 |
Hb_032107_010 |
0.2718886727 |
- |
- |
PREDICTED: uncharacterized protein LOC104225171 [Nicotiana sylvestris] |
| 16 |
Hb_000513_010 |
0.2736558802 |
- |
- |
- |
| 17 |
Hb_070268_010 |
0.2754082098 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 18 |
Hb_000232_050 |
0.2772358022 |
- |
- |
- |
| 19 |
Hb_136367_010 |
0.2824870838 |
- |
- |
hypothetical protein POPTR_0012s02040g [Populus trichocarpa] |
| 20 |
Hb_086639_060 |
0.2865953705 |
- |
- |
hypothetical protein JCGZ_25622 [Jatropha curcas] |