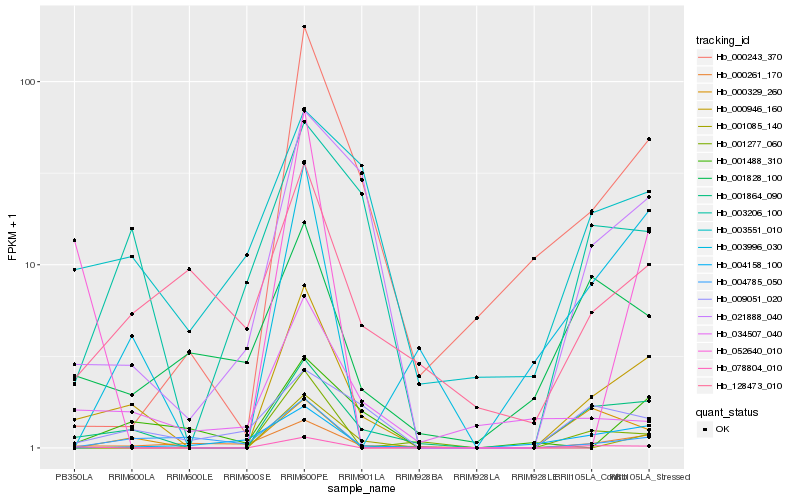
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000946_160 |
0.0 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 2 |
Hb_001864_090 |
0.1479507168 |
- |
- |
- |
| 3 |
Hb_021888_040 |
0.2816291788 |
- |
- |
PREDICTED: uncharacterized protein LOC103499290 isoform X2 [Cucumis melo] |
| 4 |
Hb_003996_030 |
0.285571025 |
- |
- |
- |
| 5 |
Hb_000243_370 |
0.2874134097 |
- |
- |
hypothetical protein VITISV_010022 [Vitis vinifera] |
| 6 |
Hb_000261_170 |
0.288400656 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 7 |
Hb_003206_100 |
0.2942465037 |
- |
- |
- |
| 8 |
Hb_078804_010 |
0.3017228803 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_004785_050 |
0.3205551383 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 10 |
Hb_001085_140 |
0.3217254795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003551_010 |
0.324535379 |
- |
- |
hypothetical protein CISIN_1g030786mg [Citrus sinensis] |
| 12 |
Hb_009051_020 |
0.3273403598 |
- |
- |
Thioredoxin O1 [Morus notabilis] |
| 13 |
Hb_001488_310 |
0.3343274314 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 14 |
Hb_128473_010 |
0.3375098769 |
- |
- |
- |
| 15 |
Hb_000329_260 |
0.3377619274 |
- |
- |
hypothetical protein MIMGU_mgv1a016162mg [Erythranthe guttata] |
| 16 |
Hb_001277_060 |
0.3387271055 |
- |
- |
ADP-ribosylation factor-like protein 5 [Morus notabilis] |
| 17 |
Hb_004158_100 |
0.3403359048 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 18 |
Hb_001828_100 |
0.3404241728 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 19 |
Hb_034507_040 |
0.3417789146 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 20 |
Hb_052640_010 |
0.3434038753 |
- |
- |
PREDICTED: putative quinone-oxidoreductase homolog, chloroplastic [Jatropha curcas] |