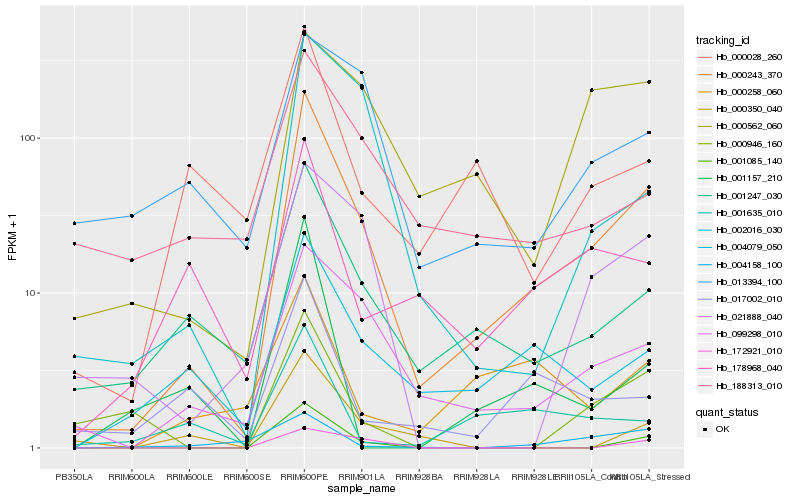
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000243_370 |
0.0 |
- |
- |
hypothetical protein VITISV_010022 [Vitis vinifera] |
| 2 |
Hb_099298_010 |
0.2165622817 |
- |
- |
- |
| 3 |
Hb_001247_030 |
0.2288896152 |
- |
- |
- |
| 4 |
Hb_004079_050 |
0.2386407331 |
- |
- |
hypothetical protein JCGZ_07229 [Jatropha curcas] |
| 5 |
Hb_001085_140 |
0.2545538271 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002016_030 |
0.2626696587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_021888_040 |
0.2656788887 |
- |
- |
PREDICTED: uncharacterized protein LOC103499290 isoform X2 [Cucumis melo] |
| 8 |
Hb_000562_060 |
0.2691547386 |
- |
- |
PREDICTED: uncharacterized protein LOC105169741 [Sesamum indicum] |
| 9 |
Hb_178968_040 |
0.2740626102 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000028_260 |
0.2763146136 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 11 |
Hb_188313_010 |
0.2787391746 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 12 |
Hb_000350_040 |
0.2872858145 |
- |
- |
- |
| 13 |
Hb_000946_160 |
0.2874134097 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 14 |
Hb_000258_060 |
0.2879314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_017002_010 |
0.2898072298 |
- |
- |
PREDICTED: expansin-A15-like [Jatropha curcas] |
| 16 |
Hb_004158_100 |
0.2962978362 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 17 |
Hb_013394_100 |
0.2964079002 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |
| 18 |
Hb_172921_010 |
0.2995070989 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |
| 19 |
Hb_001157_210 |
0.3013248674 |
- |
- |
hypothetical protein CISIN_1g038260mg [Citrus sinensis] |
| 20 |
Hb_001635_010 |
0.3014930082 |
- |
- |
- |