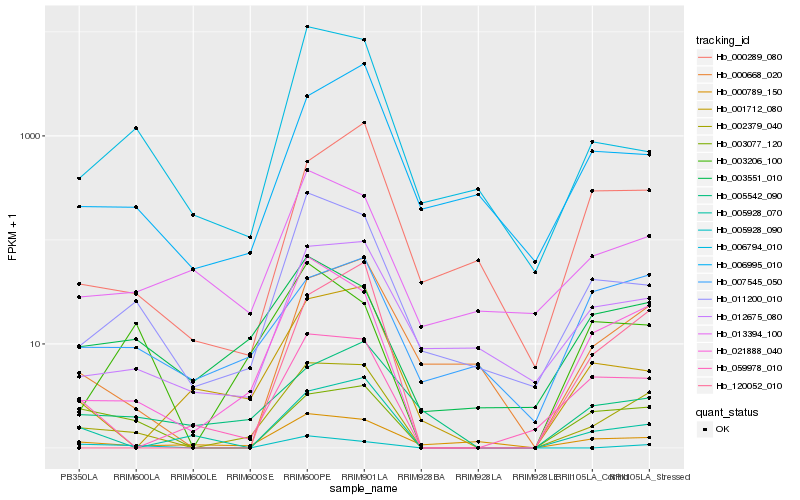
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002379_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_021888_040 |
0.1734910272 |
- |
- |
PREDICTED: uncharacterized protein LOC103499290 isoform X2 [Cucumis melo] |
| 3 |
Hb_011200_010 |
0.2203766758 |
- |
- |
- |
| 4 |
Hb_059978_010 |
0.2312488796 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 5 |
Hb_000668_020 |
0.2398264221 |
- |
- |
- |
| 6 |
Hb_012675_080 |
0.241612387 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 7 |
Hb_003551_010 |
0.2463164471 |
- |
- |
hypothetical protein CISIN_1g030786mg [Citrus sinensis] |
| 8 |
Hb_005928_070 |
0.2490357215 |
- |
- |
hypothetical protein POPTR_0002s04520g [Populus trichocarpa] |
| 9 |
Hb_013394_100 |
0.25228088 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |
| 10 |
Hb_000789_150 |
0.252397954 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_120052_010 |
0.2524867152 |
- |
- |
hypothetical protein PRUPE_ppa1027171mg [Prunus persica] |
| 12 |
Hb_001712_080 |
0.2526640593 |
- |
- |
- |
| 13 |
Hb_006794_010 |
0.2551887004 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 14 |
Hb_006995_010 |
0.2638196984 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein [Jatropha curcas] |
| 15 |
Hb_003077_120 |
0.2671512228 |
- |
- |
- |
| 16 |
Hb_000289_080 |
0.2706581038 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003206_100 |
0.2706611322 |
- |
- |
- |
| 18 |
Hb_005542_090 |
0.2710811412 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Elaeis guineensis] |
| 19 |
Hb_005928_090 |
0.2719409541 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X3 [Jatropha curcas] |
| 20 |
Hb_007545_050 |
0.2745712183 |
- |
- |
- |