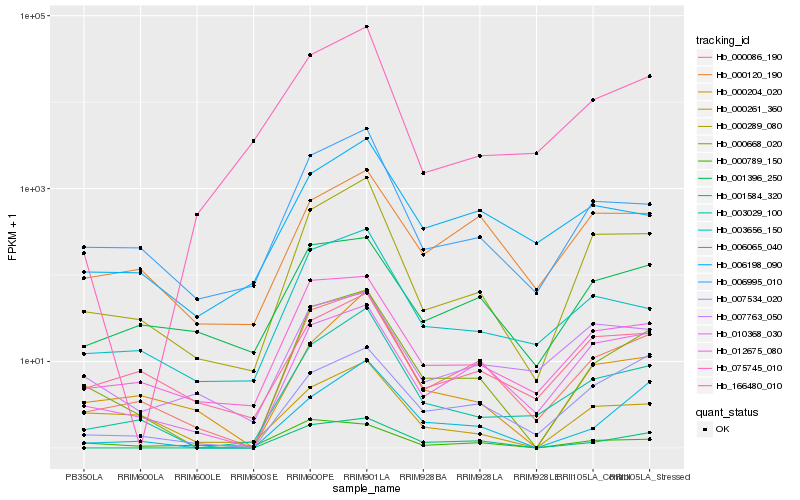
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000668_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000086_190 |
0.1169634231 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 3-like protein [Cucumis sativus] |
| 3 |
Hb_000289_080 |
0.1507946801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001584_320 |
0.1552122547 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 3 [Jatropha curcas] |
| 5 |
Hb_012675_080 |
0.1618636284 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 6 |
Hb_006995_010 |
0.1649786795 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein [Jatropha curcas] |
| 7 |
Hb_003029_100 |
0.1713641751 |
- |
- |
- |
| 8 |
Hb_010368_030 |
0.1745848591 |
- |
- |
BnaA07g35570D [Brassica napus] |
| 9 |
Hb_006065_040 |
0.1776467556 |
- |
- |
- |
| 10 |
Hb_003656_150 |
0.1909002526 |
- |
- |
- |
| 11 |
Hb_001396_250 |
0.2064627865 |
- |
- |
PREDICTED: stress-associated endoplasmic reticulum protein 2 [Jatropha curcas] |
| 12 |
Hb_000204_020 |
0.2069364766 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 13 |
Hb_000261_360 |
0.2112687436 |
- |
- |
unknown [Lotus japonicus] |
| 14 |
Hb_166480_010 |
0.2126792689 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 15 |
Hb_007534_020 |
0.2151788401 |
- |
- |
- |
| 16 |
Hb_000789_150 |
0.2162828481 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_075745_010 |
0.2203621167 |
- |
- |
hypothetical protein EUGRSUZ_D00579 [Eucalyptus grandis] |
| 18 |
Hb_006198_090 |
0.2221557582 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |
| 19 |
Hb_007763_050 |
0.225547381 |
- |
- |
hypothetical protein L484_012752 [Morus notabilis] |
| 20 |
Hb_000120_190 |
0.2284110559 |
- |
- |
Pre-mRNA-splicing factor ini1 [Ricinus communis] |