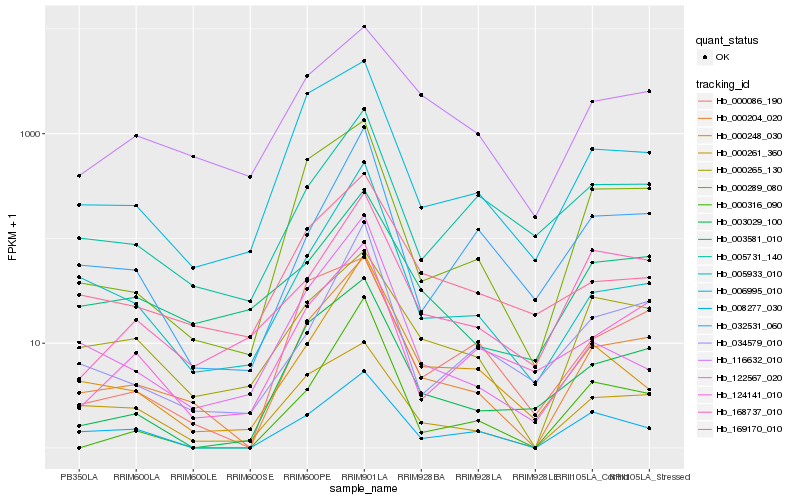
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000204_020 |
0.0 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 2 |
Hb_000289_080 |
0.1506243698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003029_100 |
0.1609149751 |
- |
- |
- |
| 4 |
Hb_006995_010 |
0.1610158642 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein [Jatropha curcas] |
| 5 |
Hb_005933_010 |
0.1718559915 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 6 |
Hb_169170_010 |
0.1760030921 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 7 |
Hb_008277_030 |
0.1819556337 |
- |
- |
hypothetical protein CICLE_v10024419mg, partial [Citrus clementina] |
| 8 |
Hb_124141_010 |
0.1829752206 |
- |
- |
plant origin recognition complex subunit, putative [Ricinus communis] |
| 9 |
Hb_000261_360 |
0.1843329376 |
- |
- |
unknown [Lotus japonicus] |
| 10 |
Hb_003581_010 |
0.1864935221 |
- |
- |
hypothetical protein VITISV_004687 [Vitis vinifera] |
| 11 |
Hb_034579_010 |
0.186500904 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000248_030 |
0.1867299253 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 13 |
Hb_168737_010 |
0.1880267232 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_116632_010 |
0.190426162 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_032531_060 |
0.192501782 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 16 |
Hb_122567_020 |
0.1947297701 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 17 |
Hb_000265_130 |
0.1956847534 |
- |
- |
- |
| 18 |
Hb_000316_090 |
0.1980477769 |
- |
- |
- |
| 19 |
Hb_005731_140 |
0.1994609332 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 20 |
Hb_000086_190 |
0.1998732636 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 3-like protein [Cucumis sativus] |