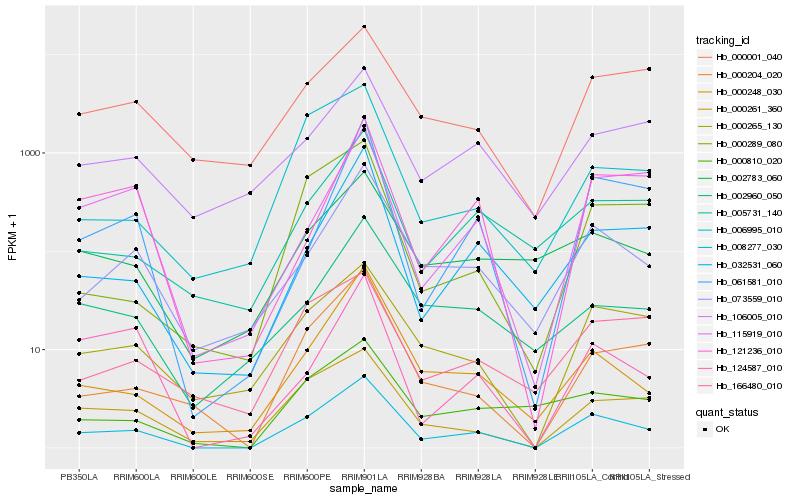
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008277_030 |
0.0 |
- |
- |
hypothetical protein CICLE_v10024419mg, partial [Citrus clementina] |
| 2 |
Hb_000265_130 |
0.1632866254 |
- |
- |
- |
| 3 |
Hb_000261_360 |
0.1660677487 |
- |
- |
unknown [Lotus japonicus] |
| 4 |
Hb_073559_010 |
0.1671306449 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 12 [Jatropha curcas] |
| 5 |
Hb_000204_020 |
0.1819556337 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 6 |
Hb_032531_060 |
0.1947035798 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 7 |
Hb_000248_030 |
0.1971684022 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 8 |
Hb_061581_010 |
0.1980074656 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_005731_140 |
0.1998940395 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 10 |
Hb_000001_040 |
0.200459955 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_121236_010 |
0.2008924161 |
- |
- |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 12 |
Hb_002783_060 |
0.2053777245 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 13 |
Hb_115919_010 |
0.205454853 |
- |
- |
unknown [Lotus japonicus] |
| 14 |
Hb_106005_010 |
0.2055497762 |
- |
- |
EG5651 [Manihot esculenta] |
| 15 |
Hb_166480_010 |
0.2060146099 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 16 |
Hb_000810_020 |
0.2072308308 |
- |
- |
- |
| 17 |
Hb_002960_050 |
0.2079410877 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 18 |
Hb_000289_080 |
0.2104361747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_006995_010 |
0.213413575 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein [Jatropha curcas] |
| 20 |
Hb_124587_010 |
0.2134584533 |
- |
- |
PREDICTED: importin-5-like [Musa acuminata subsp. malaccensis] |