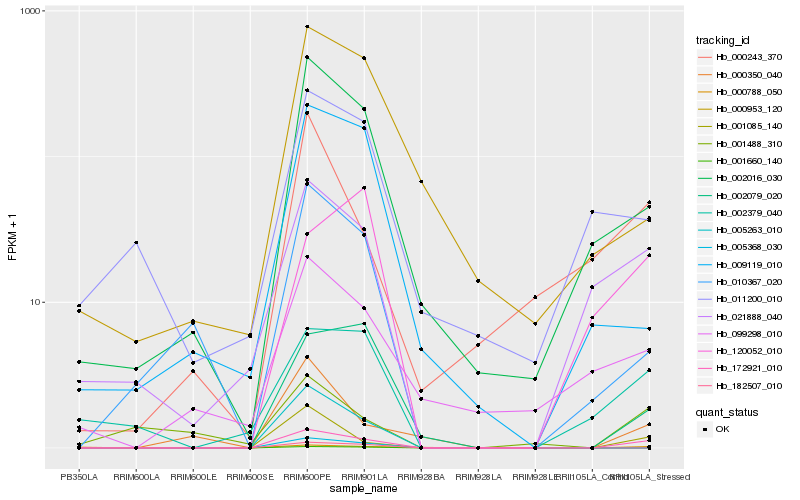
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172921_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |
| 2 |
Hb_001085_140 |
0.2186185365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002016_030 |
0.2514428512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_021888_040 |
0.2630432754 |
- |
- |
PREDICTED: uncharacterized protein LOC103499290 isoform X2 [Cucumis melo] |
| 5 |
Hb_002079_020 |
0.2733382484 |
- |
- |
- |
| 6 |
Hb_002379_040 |
0.2906250184 |
- |
- |
- |
| 7 |
Hb_000350_040 |
0.2920380954 |
- |
- |
- |
| 8 |
Hb_000243_370 |
0.2995070989 |
- |
- |
hypothetical protein VITISV_010022 [Vitis vinifera] |
| 9 |
Hb_010367_020 |
0.3036193558 |
- |
- |
hypothetical protein POPTR_0009s054801g, partial [Populus trichocarpa] |
| 10 |
Hb_000788_050 |
0.3128824988 |
- |
- |
Chloride channel protein CLC-c [Populus trichocarpa] |
| 11 |
Hb_099298_010 |
0.3203270789 |
- |
- |
- |
| 12 |
Hb_009119_010 |
0.3260015442 |
- |
- |
- |
| 13 |
Hb_000953_120 |
0.3262670332 |
- |
- |
- |
| 14 |
Hb_001488_310 |
0.3301431429 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 15 |
Hb_005368_030 |
0.330270062 |
- |
- |
- |
| 16 |
Hb_005263_010 |
0.3338036694 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Nicotiana sylvestris] |
| 17 |
Hb_120052_010 |
0.3368991703 |
- |
- |
hypothetical protein PRUPE_ppa1027171mg [Prunus persica] |
| 18 |
Hb_182507_010 |
0.33713054 |
- |
- |
- |
| 19 |
Hb_001660_140 |
0.3377909869 |
- |
- |
- |
| 20 |
Hb_011200_010 |
0.3391507649 |
- |
- |
- |