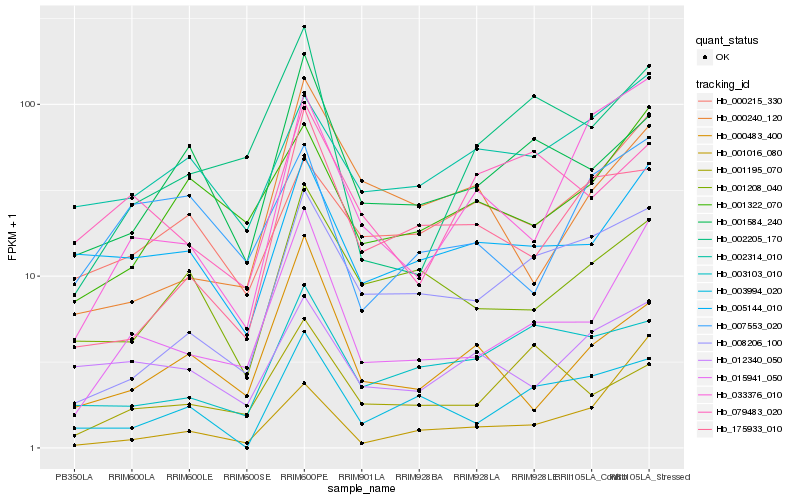
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015941_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002205_170 |
0.1841604928 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 3 |
Hb_005144_010 |
0.1989324626 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 4 |
Hb_001322_070 |
0.2012845325 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_001584_240 |
0.204643023 |
- |
- |
ADP-ribosylation factor [Gossypium barbadense] |
| 6 |
Hb_008206_100 |
0.2052082875 |
- |
- |
PREDICTED: uncharacterized protein LOC105638302 [Jatropha curcas] |
| 7 |
Hb_000483_400 |
0.2080614886 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 8 |
Hb_033376_010 |
0.2081030801 |
- |
- |
hypothetical protein POPTR_0008s02960g [Populus trichocarpa] |
| 9 |
Hb_001208_040 |
0.2179755798 |
- |
- |
hypothetical protein CISIN_1g025613mg [Citrus sinensis] |
| 10 |
Hb_000240_120 |
0.2228629546 |
- |
- |
ras-related protein Rab-5B [Reticulomyxa filosa] |
| 11 |
Hb_175933_010 |
0.2272981522 |
- |
- |
NADH-ubiquinone oxidoreductase 24 kD subunit, putative [Ricinus communis] |
| 12 |
Hb_003994_020 |
0.2310959549 |
- |
- |
- |
| 13 |
Hb_001195_070 |
0.2328928608 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 14 |
Hb_001016_080 |
0.2331954908 |
- |
- |
hypothetical protein EUGRSUZ_J02452 [Eucalyptus grandis] |
| 15 |
Hb_000215_330 |
0.2350772538 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 16 |
Hb_002314_010 |
0.2364197431 |
- |
- |
hypothetical protein JCGZ_15712 [Jatropha curcas] |
| 17 |
Hb_007553_020 |
0.2381145688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_079483_020 |
0.2381866274 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_003103_010 |
0.2383673086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_012340_050 |
0.2383733795 |
- |
- |
PREDICTED: actin-related protein 5 [Jatropha curcas] |