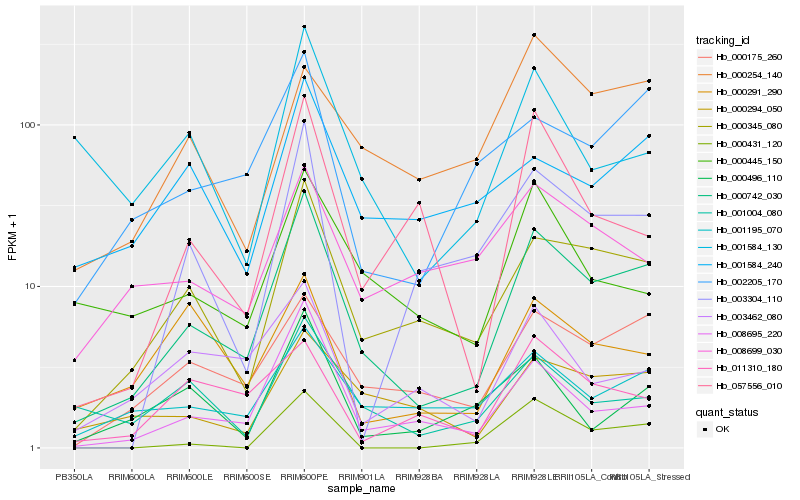
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000742_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 2 |
Hb_000345_080 |
0.1532165126 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 3 |
Hb_000175_260 |
0.1756262918 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 4 |
Hb_008695_220 |
0.1822602517 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 5 |
Hb_001195_070 |
0.1971070279 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 6 |
Hb_002205_170 |
0.1987882503 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 7 |
Hb_000294_050 |
0.201722153 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 8 |
Hb_001004_080 |
0.2182192395 |
- |
- |
hypothetical protein POPTR_0014s14130g [Populus trichocarpa] |
| 9 |
Hb_003304_110 |
0.2198144436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000445_150 |
0.226783389 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 11 |
Hb_003462_080 |
0.2268648331 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_001584_240 |
0.2290828092 |
- |
- |
ADP-ribosylation factor [Gossypium barbadense] |
| 13 |
Hb_000496_110 |
0.2295364574 |
- |
- |
PREDICTED: uncharacterized protein LOC105650214 [Jatropha curcas] |
| 14 |
Hb_000254_140 |
0.2313196832 |
- |
- |
PREDICTED: 30S ribosomal protein S17, chloroplastic [Jatropha curcas] |
| 15 |
Hb_057556_010 |
0.2316380422 |
- |
- |
Light-regulated protein precursor, putative [Ricinus communis] |
| 16 |
Hb_000431_120 |
0.232726854 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 17 |
Hb_001584_130 |
0.2335929158 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 18 |
Hb_011310_180 |
0.2343592952 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_008699_030 |
0.2344404414 |
- |
- |
hypothetical protein CISIN_1g017963mg [Citrus sinensis] |
| 20 |
Hb_000291_290 |
0.2370319916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |