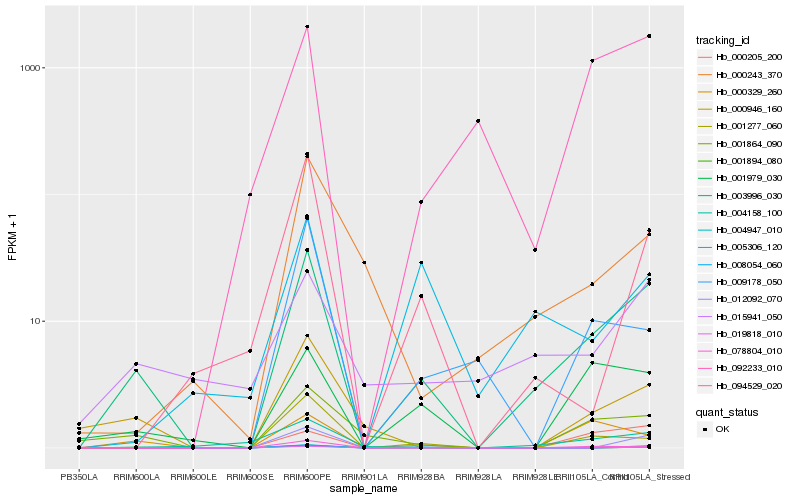
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003996_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_001979_030 |
0.2765173719 |
- |
- |
hypothetical protein POPTR_0016s06130g [Populus trichocarpa] |
| 3 |
Hb_001864_090 |
0.2780750719 |
- |
- |
- |
| 4 |
Hb_004158_100 |
0.2792197713 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 5 |
Hb_000946_160 |
0.285571025 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 6 |
Hb_015941_050 |
0.2997001661 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001277_060 |
0.3004690285 |
- |
- |
ADP-ribosylation factor-like protein 5 [Morus notabilis] |
| 8 |
Hb_001894_080 |
0.3047533487 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000205_200 |
0.3068977083 |
- |
- |
- |
| 10 |
Hb_092233_010 |
0.3089327996 |
- |
- |
RecName: Full=Profilin-3; AltName: Full=Pollen allergen Hev b 8.0201; AltName: Allergen=Hev b 8.0201 [Hevea brasiliensis] |
| 11 |
Hb_078804_010 |
0.319973783 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_094529_020 |
0.3205154748 |
- |
- |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 13 |
Hb_004947_010 |
0.3213416941 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 14 |
Hb_000329_260 |
0.3227335039 |
- |
- |
hypothetical protein MIMGU_mgv1a016162mg [Erythranthe guttata] |
| 15 |
Hb_000243_370 |
0.3268709894 |
- |
- |
hypothetical protein VITISV_010022 [Vitis vinifera] |
| 16 |
Hb_009178_050 |
0.3318306846 |
- |
- |
Elongation factor 1-alpha, putative [Ricinus communis] |
| 17 |
Hb_005306_120 |
0.3334065536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_008054_060 |
0.3373521608 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Populus euphratica] |
| 19 |
Hb_012092_070 |
0.3374764696 |
- |
- |
PREDICTED: uncharacterized protein LOC105644084 [Jatropha curcas] |
| 20 |
Hb_019818_010 |
0.3377626118 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |