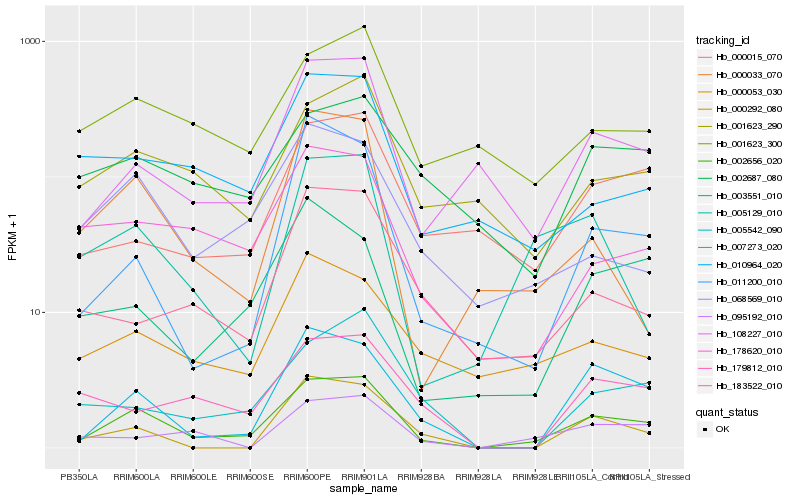
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002656_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_007273_020 |
0.1742622762 |
- |
- |
PREDICTED: probable U6 snRNA-associated Sm-like protein LSm4 [Cicer arietinum] |
| 3 |
Hb_108227_010 |
0.1968597023 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 4 |
Hb_178620_010 |
0.2105907778 |
- |
- |
ADP-ribosylation factor, arf, putative [Ricinus communis] |
| 5 |
Hb_068569_010 |
0.2114660184 |
- |
- |
- |
| 6 |
Hb_001623_290 |
0.2129285958 |
- |
- |
unknown [Populus trichocarpa] |
| 7 |
Hb_003551_010 |
0.2182318189 |
- |
- |
hypothetical protein CISIN_1g030786mg [Citrus sinensis] |
| 8 |
Hb_000053_030 |
0.2184601845 |
- |
- |
hypothetical protein CISIN_1g0010393mg, partial [Citrus sinensis] |
| 9 |
Hb_001623_300 |
0.2194941652 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-B [Jatropha curcas] |
| 10 |
Hb_005542_090 |
0.2203669496 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Elaeis guineensis] |
| 11 |
Hb_000015_070 |
0.223753941 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |
| 12 |
Hb_095192_010 |
0.2279033956 |
- |
- |
hypothetical protein VITISV_043980 [Vitis vinifera] |
| 13 |
Hb_011200_010 |
0.2289895926 |
- |
- |
- |
| 14 |
Hb_010964_020 |
0.2290237774 |
- |
- |
RNA helicase [Arabidopsis thaliana] |
| 15 |
Hb_183522_010 |
0.2297984221 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 16 |
Hb_179812_010 |
0.2302038715 |
- |
- |
TdcA1-ORF1-ORF2 [Daucus carota] |
| 17 |
Hb_005129_010 |
0.2319245737 |
- |
- |
hypothetical protein VITISV_026364 [Vitis vinifera] |
| 18 |
Hb_000292_080 |
0.2321017116 |
- |
- |
hypothetical protein CISIN_1g034193mg [Citrus sinensis] |
| 19 |
Hb_002687_080 |
0.2321040135 |
- |
- |
hypothetical protein JCGZ_06535 [Jatropha curcas] |
| 20 |
Hb_000033_070 |
0.2343492196 |
- |
- |
unnamed protein product [Oryza sativa Japonica Group] |