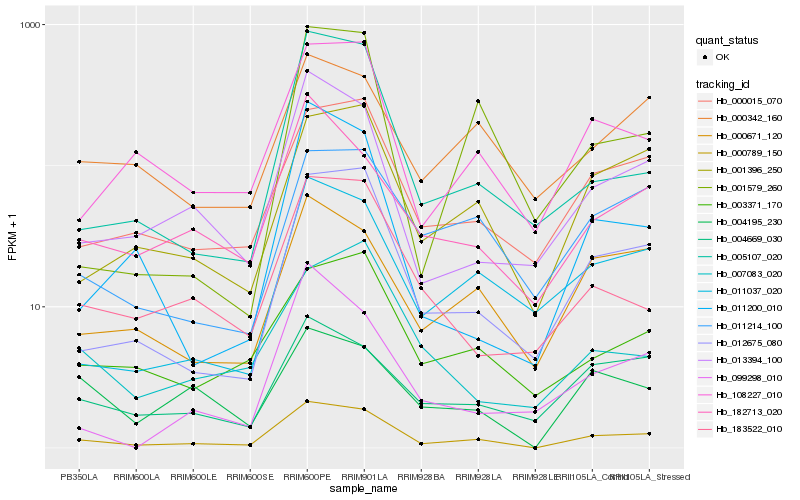
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000789_150 |
0.0 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_012675_080 |
0.1514236991 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 3 |
Hb_013394_100 |
0.1580749357 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |
| 4 |
Hb_108227_010 |
0.1620860647 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 5 |
Hb_000015_070 |
0.1665137257 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |
| 6 |
Hb_001396_250 |
0.169481065 |
- |
- |
PREDICTED: stress-associated endoplasmic reticulum protein 2 [Jatropha curcas] |
| 7 |
Hb_000671_120 |
0.1739934159 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 8 |
Hb_005107_020 |
0.175506423 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 9 |
Hb_182713_020 |
0.1779066828 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727 [Setaria italica] |
| 10 |
Hb_011037_020 |
0.1781988105 |
- |
- |
unknown [Picea sitchensis] |
| 11 |
Hb_003371_170 |
0.1829591977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_183522_010 |
0.1836681407 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 13 |
Hb_011214_100 |
0.1931872677 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 14 |
Hb_004669_030 |
0.1937923558 |
- |
- |
- |
| 15 |
Hb_004195_230 |
0.1955254562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011200_010 |
0.196399922 |
- |
- |
- |
| 17 |
Hb_000342_160 |
0.2033778318 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 18 |
Hb_007083_020 |
0.2102078919 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |
| 19 |
Hb_001579_260 |
0.2119613223 |
- |
- |
putative ADP-ribolylation factor [Arabidopsis thaliana] |
| 20 |
Hb_099298_010 |
0.2136609469 |
- |
- |
- |