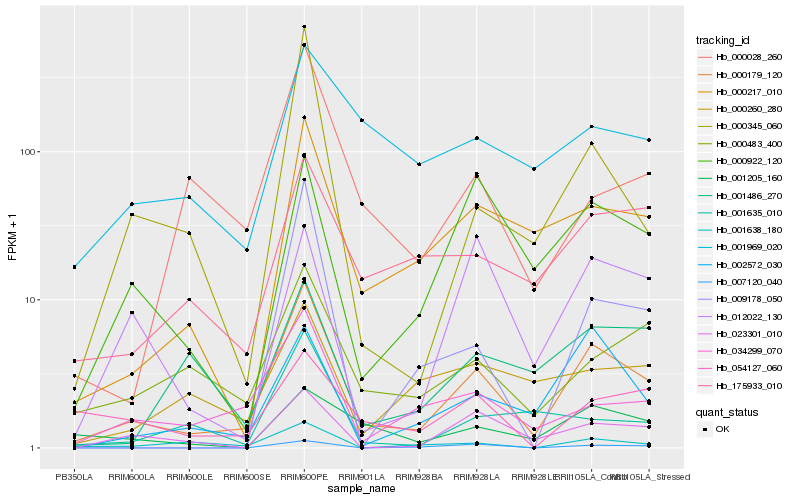
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000179_120 |
0.0 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 2 |
Hb_000217_010 |
0.2193759352 |
- |
- |
hypothetical protein POPTR_0341s00210g [Populus trichocarpa] |
| 3 |
Hb_034299_070 |
0.226241307 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 4 |
Hb_000345_060 |
0.2320371844 |
- |
- |
PREDICTED: REF/SRPP-like protein At3g05500 [Jatropha curcas] |
| 5 |
Hb_000483_400 |
0.2387323934 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 6 |
Hb_002572_030 |
0.2423688018 |
- |
- |
PREDICTED: diacylglycerol kinase theta [Jatropha curcas] |
| 7 |
Hb_023301_010 |
0.2434071134 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |
| 8 |
Hb_001638_180 |
0.2441374645 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 9 |
Hb_175933_010 |
0.2502250658 |
- |
- |
NADH-ubiquinone oxidoreductase 24 kD subunit, putative [Ricinus communis] |
| 10 |
Hb_000922_120 |
0.2506078351 |
- |
- |
PREDICTED: carbonic anhydrase 2-like [Jatropha curcas] |
| 11 |
Hb_000028_260 |
0.251709823 |
- |
- |
hypothetical protein glysoja_041948 [Glycine soja] |
| 12 |
Hb_009178_050 |
0.2643762693 |
- |
- |
Elongation factor 1-alpha, putative [Ricinus communis] |
| 13 |
Hb_001635_010 |
0.2647300771 |
- |
- |
- |
| 14 |
Hb_007120_040 |
0.2675233457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001205_160 |
0.2705183443 |
- |
- |
hypothetical protein POPTR_0005s12560g [Populus trichocarpa] |
| 16 |
Hb_054127_060 |
0.2717507866 |
- |
- |
hypothetical protein POPTR_0004s09330g [Populus trichocarpa] |
| 17 |
Hb_012022_130 |
0.273974654 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 18 |
Hb_001486_270 |
0.2747855271 |
- |
- |
Ann10 [Manihot esculenta] |
| 19 |
Hb_001969_020 |
0.275153169 |
- |
- |
hypothetical protein POPTR_0010s04700g [Populus trichocarpa] |
| 20 |
Hb_000260_280 |
0.2761649756 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |