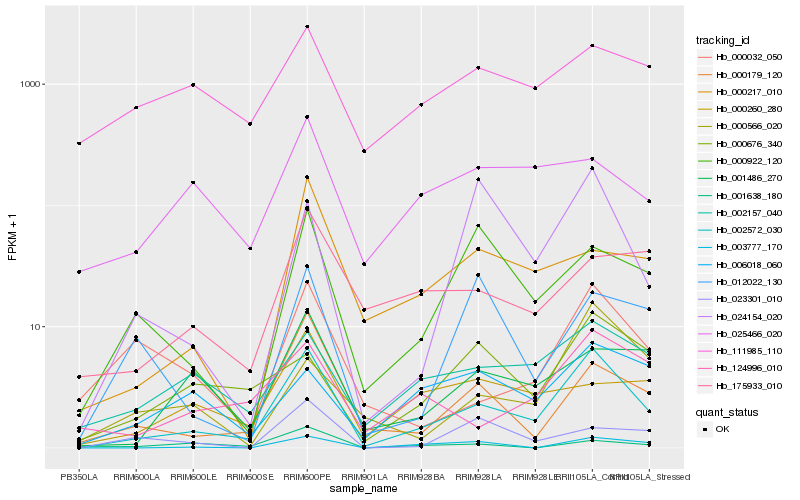
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002572_030 |
0.0 |
- |
- |
PREDICTED: diacylglycerol kinase theta [Jatropha curcas] |
| 2 |
Hb_000179_120 |
0.2423688018 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 3 |
Hb_002157_040 |
0.2603978987 |
- |
- |
PREDICTED: uncharacterized protein LOC105628778 [Jatropha curcas] |
| 4 |
Hb_001486_270 |
0.2628243006 |
- |
- |
Ann10 [Manihot esculenta] |
| 5 |
Hb_003777_170 |
0.2628824088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_124996_010 |
0.2635213812 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 7 |
Hb_000032_050 |
0.266330091 |
- |
- |
hypothetical protein RCOM_1691130 [Ricinus communis] |
| 8 |
Hb_000922_120 |
0.2756556673 |
- |
- |
PREDICTED: carbonic anhydrase 2-like [Jatropha curcas] |
| 9 |
Hb_000217_010 |
0.2842016026 |
- |
- |
hypothetical protein POPTR_0341s00210g [Populus trichocarpa] |
| 10 |
Hb_000566_020 |
0.2864278001 |
- |
- |
hypothetical protein POPTR_0001s13520g [Populus trichocarpa] |
| 11 |
Hb_024154_020 |
0.2868701314 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 12 |
Hb_000676_340 |
0.2899794152 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_175933_010 |
0.291286524 |
- |
- |
NADH-ubiquinone oxidoreductase 24 kD subunit, putative [Ricinus communis] |
| 14 |
Hb_000260_280 |
0.2922074265 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 15 |
Hb_006018_060 |
0.2971545543 |
- |
- |
Zinc finger SWIM domain-containing protein 7 isoform 1 [Theobroma cacao] |
| 16 |
Hb_025466_020 |
0.3024873963 |
- |
- |
tubulin beta-6 [Brassica oleracea var. italica] |
| 17 |
Hb_023301_010 |
0.3054000588 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |
| 18 |
Hb_111985_110 |
0.3079110486 |
- |
- |
thioredoxin H-type 3 [Hevea brasiliensis] |
| 19 |
Hb_001638_180 |
0.3079224575 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 20 |
Hb_012022_130 |
0.3111477993 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |